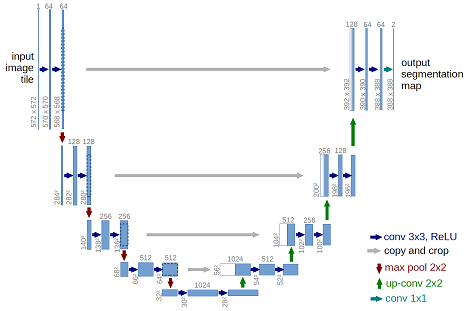
The architecture was inspired by U-Net: Convolutional Networks for Biomedical Image Segmentation.
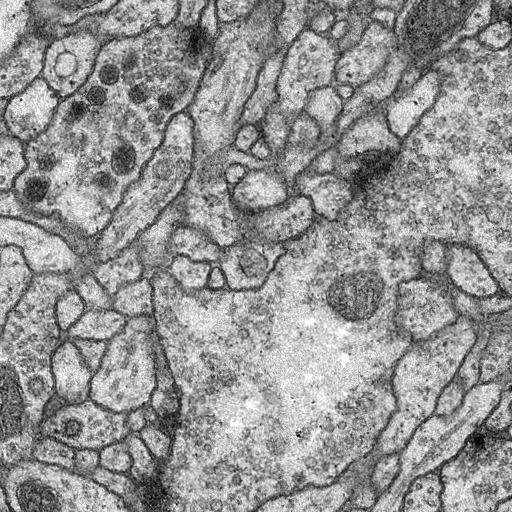
The original dataset is from isbi challenge, and I've downloaded it and done the pre-processing.
You can find it in folder data/membrane.
The data for training contains 30 512*512 images, which are far not enough to feed a deep learning neural network. I use a module called ImageDataGenerator in keras.preprocessing.image to do data augmentation.
See dataPrepare.ipynb and data.py for detail.
This deep neural network is implemented with Keras functional API, which makes it extremely easy to experiment with different interesting architectures.
Output from the network is a 512*512 which represents mask that should be learned. Sigmoid activation function makes sure that mask pixels are in [0, 1] range.
The model is trained for 5 epochs.
After 5 epochs, calculated accuracy is about 0.97.
Loss function for the training is basically just a binary crossentropy.
This tutorial depends on the following libraries:
- Tensorflow
- Keras >= 1.0
Also, this code should be compatible with Python versions 2.7-3.5.
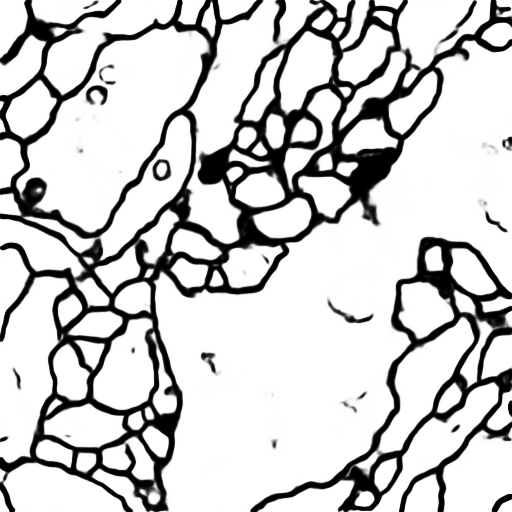
You will see the predicted results of test image in data/membrane/test
Use the trained model to do segmentation on test images, the result is statisfactory.
Keras is a minimalist, highly modular neural networks library, written in Python and capable of running on top of either TensorFlow or Theano. It was developed with a focus on enabling fast experimentation. Being able to go from idea to result with the least possible delay is key to doing good research.
Use Keras if you need a deep learning library that:
allows for easy and fast prototyping (through total modularity, minimalism, and extensibility). supports both convolutional networks and recurrent networks, as well as combinations of the two. supports arbitrary connectivity schemes (including multi-input and multi-output training). runs seamlessly on CPU and GPU. Read the documentation Keras.io
Keras is compatible with: Python 2.7-3.5.