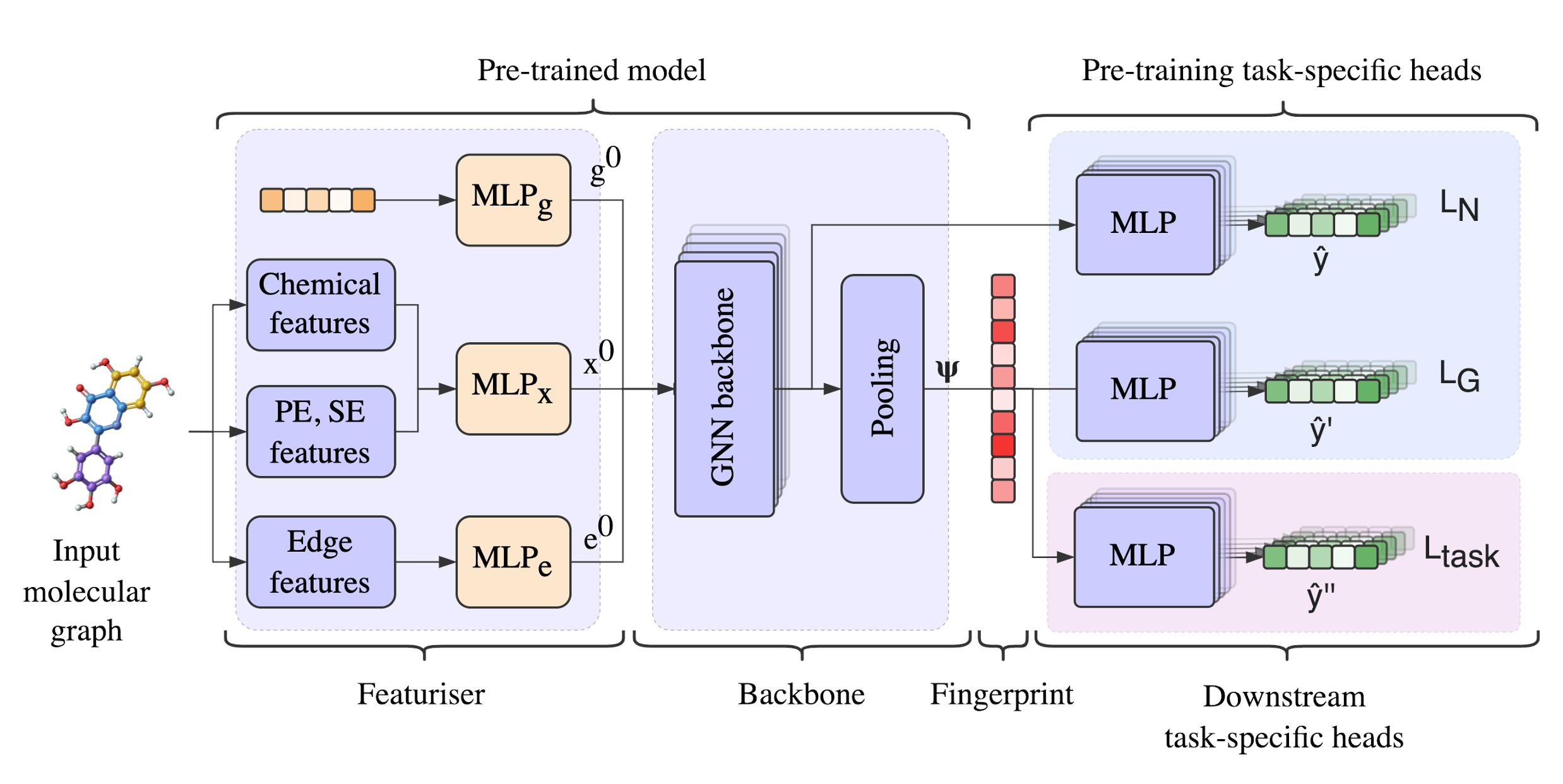
A parameter-efficient molecular featuriser that generalises well to biological tasks thanks to the effective pre-training on biological and quantum mechnical datasets.
The model has been introduced in the paper 𝙼𝚒𝚗𝚒𝙼𝚘𝚕: A Parameter-Efficient Foundation Model for Molecular Learning, published in the ICML workshop on Accessible and Efficient Foundation Models for Biological Discovery in 2024.
Embeddings can be generated in four lines of code:
from minimol import Minimol
model = Minimol()
smiles = [
'COc1ccc2cc(C(=O)NC3(C(=O)N[C@H](Cc4ccccc4)C(=O)NCC4CCN(CC5CCOCC5)CC4)CCCC3)sc2c1',
'Nc1nc(=O)c2c([nH]1)NCC(CNc1ccc(C(=O)NC(CCC(=O)O)C(=O)O)cc1)N2C=O',
'O=C1CCCN1CCCCN1CCN(c2cc(C(F)(F)F)ccn2)CC1',
'c1ccc(-c2cccnc2)cc1',
]
model(smiles)
>> A list of 4 tensors of (512,) shape
For a Colab notebook showing how to use Minimol's fingerprints to achieve SoTA results on a downstream task, click here:
When used with cuda, use nvcc --version to see which version of the driver is installed on your machine, to select the wheel (cuXXX):
pip install torch-sparse torch-cluster torch-scatter -f https://pytorch-geometric.com/whl/torch-2.3.0+cu124.html
pip install minimol
git clone git@github.com:graphcore-research/minimol.git
cd minimol
mamba env create -f env.yml -n minimol_venv
mamba activate minimol
To install mamba see the official documentation.
The model has been evaluated on 22 benchmarks from the ADMET group of Therapeutics Data Commons (TDC). These are the results when comparing to MolE and TOP5 models from the TDC leaderboard (as of June 2024):
| TDC Dataset | TDC Leaderboard | MolE | MiniMol (GINE) | ||||
|---|---|---|---|---|---|---|---|
| Name | Size | Metric | SoTA Result | Result | Rank | Result | Rank |
| Absorption | |||||||
| Caco2 Wang | 906 | MAE | 0.276 ± 0.005 | 0.310 ± 0.010 | 6 | 0.350 ± 0.018 | 7 |
| Bioavailability Ma | 640 | AUROC | 0.748 ± 0.033 | 0.654 ± 0.028 | 7 | 0.689 ± 0.020 | 5 |
| Lipophilicity AZ | 4,200 | MAE | 0.467 ± 0.006 | 0.469 ± 0.009 | 3 | 0.456 ± 0.008 | 1 |
| Solubility AqSolDB | 9,982 | MAE | 0.761 ± 0.025 | 0.792 ± 0.005 | 5 | 0.741 ± 0.013 | 1 |
| HIA Hou | 578 | AUROC | 0.989 ± 0.001 | 0.963 ± 0.019 | 7 | 0.993 ± 0.005 | 1 |
| Pgp Broccatelli | 1,212 | AUROC | 0.938 ± 0.002 | 0.915 ± 0.005 | 7 | 0.942 ± 0.002 | 1 |
| Distribution | |||||||
| BBB Martins | 1,975 | AUROC | 0.916 ± 0.001 | 0.903 ± 0.005 | 7 | 0.924 ± 0.003 | 1 |
| PPBR AZ | 1,797 | MAE | 7.526 ± 0.106 | 8.073 ± 0.335 | 6 | 7.696 ± 0.125 | 4 |
| VDss Lombardo | 1,130 | Spearman | 0.713 ± 0.007 | 0.654 ± 0.031 | 3 | 0.535 ± 0.027 | 7 |
| Metabolism | |||||||
| CYP2C9 Veith | 12,092 | AUPRC | 0.859 ± 0.001 | 0.801 ± 0.003 | 5 | 0.823 ± 0.006 | 4 |
| CYP2D6 Veith | 13,130 | AUPRC | 0.790 ± 0.001 | 0.682 ± 0.008 | 6 | 0.719 ± 0.004 | 5 |
| CYP3A4 Veith | 12,328 | AUPRC | 0.916 ± 0.000 | 0.867 ± 0.003 | 7 | 0.877 ± 0.001 | 4 |
| CYP2C9 Substrate | 666 | AUPRC | 0.441 ± 0.033 | 0.446 ± 0.062 | 2 | 0.474 ± 0.025 | 1 |
| CYP2D6 Substrate | 664 | AUPRC | 0.736 ± 0.024 | 0.699 ± 0.018 | 7 | 0.695 ± 0.032 | 6 |
| CYP3A4 Substrate | 667 | AUROC | 0.662 ± 0.031 | 0.670 ± 0.018 | 1 | 0.663 ± 0.008 | 2 |
| Excretion | |||||||
| Half Life Obach | 667 | Spearman | 0.562 ± 0.008 | 0.549 ± 0.024 | 4 | 0.495 ± 0.042 | 6 |
| Clearance Hepatocyte | 1,102 | Spearman | 0.498 ± 0.009 | 0.381 ± 0.038 | 7 | 0.446 ± 0.029 | 3 |
| Clearance Microsome | 1,020 | Spearman | 0.630 ± 0.010 | 0.607 ± 0.027 | 6 | 0.628 ± 0.005 | 2 |
| Toxicity | |||||||
| LD50 Zhu | 7,385 | MAE | 0.552 ± 0.009 | 0.823 ± 0.019 | 7 | 0.585 ± 0.005 | 2 |
| hERG | 648 | AUROC | 0.880 ± 0.002 | 0.813 ± 0.009 | 7 | 0.846 ± 0.016 | 4 |
| Ames | 7,255 | AUROC | 0.871 ± 0.002 | 0.883 ± 0.005 | 1 | 0.849 ± 0.004 | 5 |
| DILI | 475 | AUROC | 0.925 ± 0.005 | 0.577 ± 0.021 | 7 | 0.956 ± 0.006 | 1 |
| Mean Rank: | 5.2 | 3.3 |
Copyright (c) 2024 Graphcore Ltd. Licensed under the MIT License.
The included code is released under the MIT license (see details of the license).