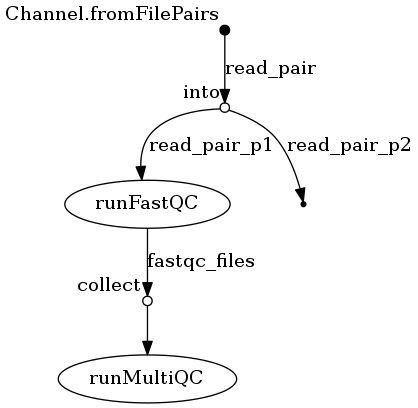
A Nextflow workflow that runs FastQC on Fastq reads and then combines them into a MultiQC report. FastQC and MutliQC are setup on a Docker container. The nextflow script pulls the Docker container and converts it to a Singularity container on the fly. See the nextflow.log for settings to run on a Slurm cluster.
To be able to mount the /data directory on the cluster you need to have the mount defined in your Docker/Singularity container e.g. see
In the nextflow.config two possible ways to pull the container are suggested. Directly from quay.io or directly from a file on the share.
To be able to mount the non native singularity conainer mounts e.g. data singularity.autoMounts = true should be set.
nextflow -log nextflow.log run -w /data/users/cbio-ws-u24/nextflow-workdir -c nextflow.config main.nf -profile slurm
$ squeue
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
2324 main tl.rFQC. cbio-ws- R 0:12 1 cbio-ws-1
2325 main tl.rFQC. cbio-ws- R 0:12 1 cbio-ws-1
2327 main tl.rFQC. cbio-ws- R 0:12 1 cbio-ws-1
2329 main tl.rFQC. cbio-ws- R 0:12 1 cbio-ws-1
2336 main tl.rFQC. cbio-ws- R 0:12 1 cbio-ws-1
srun -N 1 --ntasks-per-node=1 --pty bash
singularity exec -B /data:/data /data/projects/cbio-ws/tests/containers/quay.io_h3abionet_org_h3a16s-fastqc.img bash
fastqc /data/projects/cbio-ws/tests/fastq-reads/Dog1_R1.fastq -o /data/users/cbio-ws-u24/
exit
ls /data/users/cbio-ws-u24/Dog1_R1_fastqc.*
/data/users/cbio-ws-u24/Dog1_R1_fastqc.html /data/users/cbio-ws-u24/Dog1_R1_fastqc.zip