Continuous Analysis Phylogentics Example
This is a sample repository showing the Continuous Analysis Workflow. This process is described in detail and available as a pre-print.
In this example we use several tools to build phylogenies from alignments:
Sample Results
The full analysis is described below, here we show two of the useful artifacts generated through Continuous Analysis:
- Change logs/synchronization between code and figures: Live Version
- Complete "audit" logs of the code run: Logs
Description of analysis
In this analysis we align 5 mRNA sequences and use these alignments to build phylogenies. Analysis code is available here
- We look at 5 mRNA sequences (findable at: http://www.ncbi.nlm.nih.gov/nuccore)
- Twist - Fly (NM_079092, splice form A)
- Twist1 - Human (NM_000474)
- Twist1 - Mouse (NM_011658)
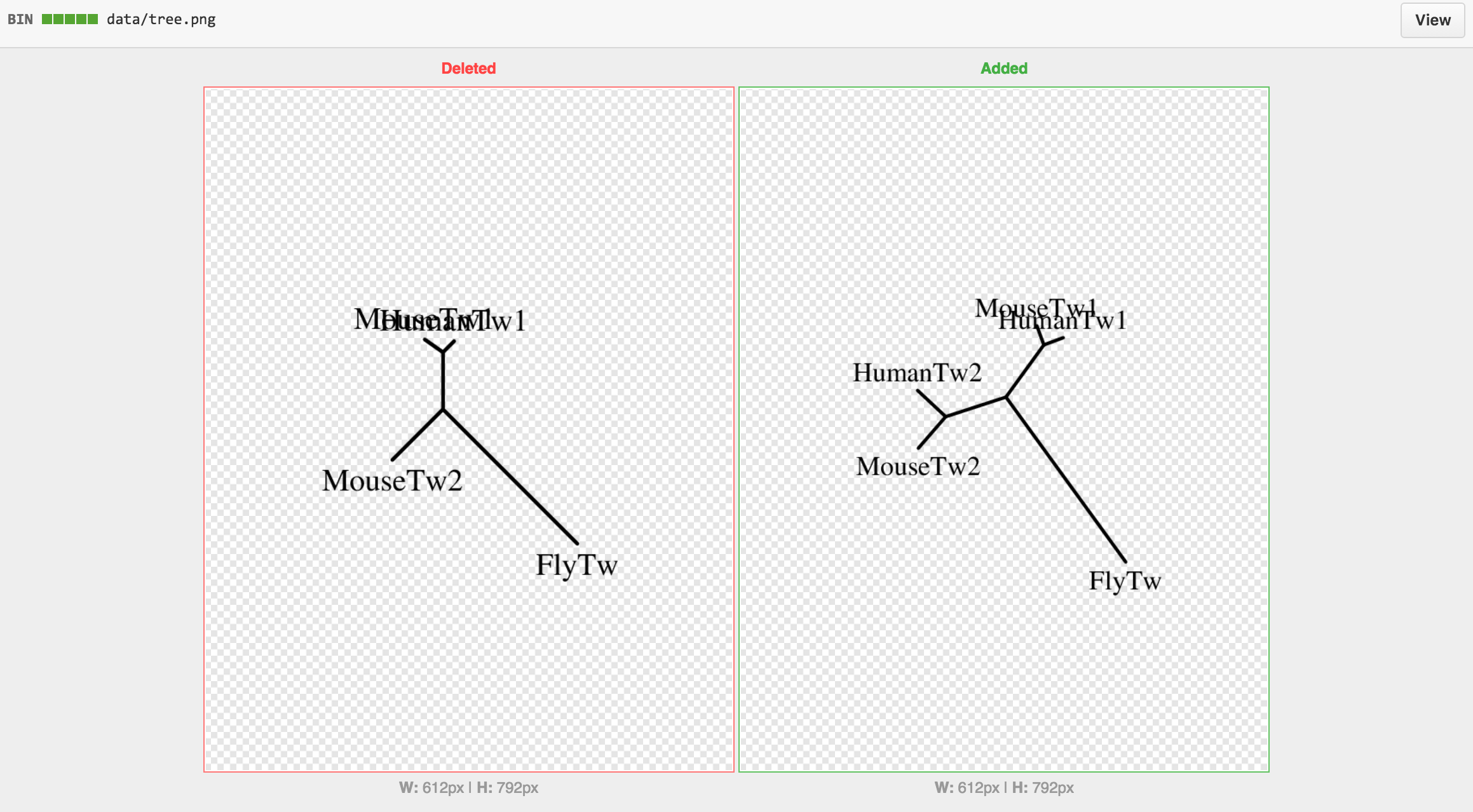
- Twist2 - Human (NM_057179) - Added in second commit to see differences
- Twist2 - Mouse (NM_007855)
And load the sequences into twist.fasta
- Align the sequences using MAFFT.
- Convert to PHYLIP interleaved format using EMBOSS Seqret.
- Calculate the maximum parsimony tree for the sequences using DNAPARS.
- Draw a representation fo this tree using drawtree.
- Use Seqboot to assess the robustness of the generated tree.
- Determine the consensus tree from the bootstrapped trees.
We performed this process twice, once without HumanTwist2 and once with HumanTwist2. The difference is viewable at: results.
Feedback
Please feel free to email me - (brettbe) at med.upenn.edu with any feedback or raise a github issue with any comments or questions.
Acknowledgements
We would like to thank Katie Siewert for providing the analysis design.
This work is supported by the Gordon and Betty Moore Foundation's Data-Driven Discovery Initiative through Grant GBMF4552 to C.S.G. as well as the Commonwealth Universal Research Enhancement (CURE) Program grant from the Pennsylvania Department of Health.