When you code C/C++ applications for DNA, you often store specific values in arrays where the index is the nucleotide or you need a numerical value for a nucleotide. A lot of people use the following:
A => 0
C => 1
G => 2
T => 3
I saw an interesting tweet about a trick to do this in a single line:
0b11 & (bp >> 2 ^ bp >> 1)
This works even for lower case DNA bases. What does this line do? It bit-shifts the numeric value of the base by 1 to the right, then 2 to the right, computes the XOR of both and uses the last 2 bits. Why does this work? Each character has an ASCII value:
A 65
C 67
G 71
T 84
a 97
c 99
g 103
t 116
Let's look at the binary value for the upper case characters:
A 1000001
C 1000011
G 1000111
T 1010100
Let's bitshift by 1 to the right:
A 0100000
C 0100001
G 0100011
T 0101010
The last 2 bits are almost there, we have 00 for A, 01 for C but 11 for G and 10 for T. Let's bit shift again:
A 0010000
C 0010000
G 0010001
T 0010101
This is interesting because we have a 0 for A,C and a 1 for G,T. If we XOR this with the first bitshift we get:
A 0110000
C 0110001
G 0110010
T 0111111
We are almost done, the last 2 bits are exactly what we need so let's mask with 3 or 11 in binary:
A 0000000
C 0000001
G 0000010
T 0000011
and we are done! How fast is this? Let's generate 10M bases at random and compute the index:
` for(int i=0;i<10000000;i++){ char bp="ACGT"[ rand()%4 ]; int idx = (0b11 & (bp >> 2 ^ bp >> 1) );
cout<<bp<<"\t"<<idx<<endl;
}
`
I coded 4 programs to do this:
- bit operators
- set of 'if' statements
- a C++ map
- an array using chars as indices
The last one simply stores the indices in an array:
char dna2int ['T'+1];
dna2int['A']=0;
dna2int['C']=1;
dna2int['G']=2;
dna2int['T']=3;
for(int i=0;i<10000000;i++){
char bp="ACGT"[ rand()%4 ];
int idx = dna2int[bp];
cout<<bp<<"\t"<<idx<<endl;
}
` I conducted 10k replicates for each. First, they all gave the same result:
./DNA2INT_array |md5sum
#6e9da6e17fb64b5eb8470cfb46e9f202 -
./DNA2INT_bitshift |md5sum
#6e9da6e17fb64b5eb8470cfb46e9f202 -
./DNA2INT_if |md5sum
#6e9da6e17fb64b5eb8470cfb46e9f202 -
./DNA2INT_map |md5sum
#6e9da6e17fb64b5eb8470cfb46e9f202 -
Here are the average runtimes:
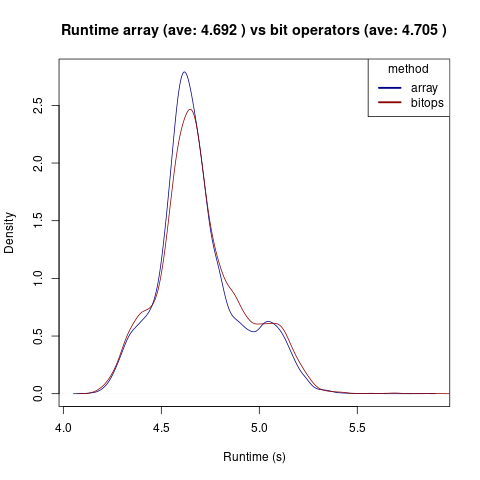
- bit operators 4.704s
- 'if' statements 5.243s
- C++ map 5.278s
- simple array 4.692s
'If' statements and a map are the slowest, they take about 5.3s on average. Bit operators were indeed much faster at 4.7s. However, a simple array was about as fast as bitwise operators with an average of 4.69s. However, the distribution of runtimes between an array and bitoperators look quite similar:
For now, arrays are still the fastest. I did not test lower case characters. It would be interesting to find an even faster solution. Also, this is not a real life scenario where there could be a cache miss resulting in higher runtimes.