An open framework for evaluating correspondence in brain images and teaching neuroanatomy using anatomical fiducial placement
Documentation: https://afidsproject.com
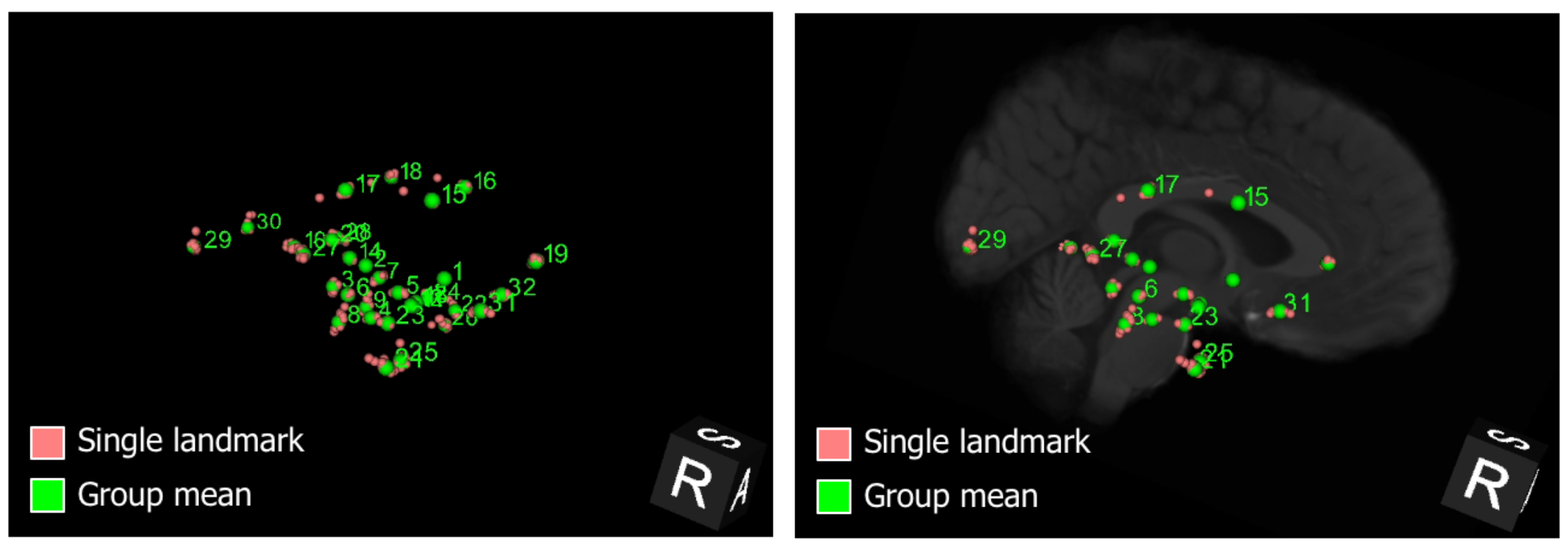
- The AFIDs protocol is an anatomical fiducial placement protocol that has been validated and used for teaching at a number of local and global BrainHack related events.
- The protocol and study framework leverage open resources/tools and has been developed with full transparency in mind so that others may freely use, adopt, and modify.
- AFID placement is reproducible, not overtly manually intensive (20-40 minutes once trained), and more sensitive to local registration errors than standard voxel overlap measures.
- 60+ raters trained to date.
- Convert3D tool: https://sourceforge.net/projects/c3d/
- further documentation for c3d: https://github.com/pyushkevich/c3d/blob/master/doc/c3d.md
- python requirements (defined in
requirements.txt):- yYAML>=5.3.1
- scikit-learn>=0.23.1
- nibabel>=3.1.0
- pandas>=1.0.5
- numpy>=1.19.4
The input directory with *_T1w.nii.gz and AFIDs .fcsv files should be setup as follows:
input_data/
├── sub-<###>_*.fcsv
├── sub-<###>_*T1w.nii.gz
└── ....fcsvis the 3D Slicer markups file with all 32 AFIDs defined for the relative T1w imageT1w.nii.gzis the T1w nifti file associated with the AFIDs markups
Edit the config/config.yaml file to include the paths to the following:
-
Clone a copy of this repository to your system:
git clone https://github.com/greydongilmore/autofids-brainhack2020.git
-
Install the Python dependencies by opening a terminal, changing to the project directory and running:
python -m pip install -r requirements.txt
All the following commands should be run in the root of the project directory.
-
Convert source
.fcsvfiles into a 3D mask using c3d:python /workflow/scripts/fcsvToLandmarks.py
-
Running the training pipeline:
python /workflow/scripts/c3dTrain.py
-
Running the testing pipeline:
python /workflow/scripts/c3dTest.py