| Package | Status | Usage | GitHub | References |
|---|---|---|---|---|
 |
 |
|||
 |
||||
 |
 |
 |
 |
|
 |
 |
|||
 |
 |
 |
||
 |
 |
 |
||
 |
pairwiseComparisons
provides a tidy data friendly way to carry out pairwise comparison
tests.
It currently supports post hoc multiple pairwise comparisons tests for both between-subjects and within-subjects one-way analysis of variance designs. For both of these designs, parametric, non-parametric, and robust statistical tests are available.
To get the latest, stable CRAN release (0.1.1):
install.packages(pkgs = "pairwiseComparisons")You can get the development version of the package from GitHub
(0.1.1.9000). To see what new changes (and bug fixes) have been made
to the package since the last release on CRAN, you can check the
detailed log of changes here:
https://indrajeetpatil.github.io/pairwiseComparisons/news/index.html
If you are in hurry and want to reduce the time of installation, prefer-
# needed package to download from GitHub repo
install.packages(pkgs = "remotes")
# downloading the package from GitHub
remotes::install_github(
repo = "IndrajeetPatil/pairwiseComparisons", # package path on GitHub
dependencies = FALSE, # assumes you have already installed needed packages
quick = TRUE # skips docs, demos, and vignettes
)If time is not a constraint-
remotes::install_github(
repo = "IndrajeetPatil/pairwiseComparisons", # package path on GitHub
dependencies = TRUE, # installs packages which pairwiseComparisons depends on
upgrade_dependencies = TRUE # updates any out of date dependencies
)Following table contains a brief summary of the currently supported pairwise comparison tests-
| Type | Equal variance? | Test | p-value adjustment? |
|---|---|---|---|
| Parametric | No | Games-Howell test | Yes |
| Parametric | Yes | Student’s t-test | Yes |
| Non-parametric | No | Dwass-Steel-Crichtlow-Fligner test | Yes |
| Robust | No | Yuen’s trimmed means test | Yes |
| Bayes Factor | No | No | No |
| Bayes Factor | Yes | No | No |
| Type | Test | p-value adjustment? |
|---|---|---|
| Parametric | Student’s t-test | Yes |
| Non-parametric | Durbin-Conover test | Yes |
| Robust | Yuen’s trimmed means test | Yes |
| Bayes Factor | No | No |
Here we will see specific examples of how to use this function for different types of
- designs (between or within subjects)
- statistics (parametric, non-parametric, robust)
- p-value adjustment methods
# for reproducibility
set.seed(123)
library(pairwiseComparisons)
# parametric
# if `var.equal = TRUE`, then Student's *t*-test will be run
pairwise_comparisons(
data = ggplot2::msleep,
x = vore,
y = brainwt,
type = "parametric",
var.equal = TRUE,
paired = FALSE,
p.adjust.method = "bonferroni"
)
#> Note: The parametric pairwise multiple comparisons test used-
#> Student's t-test.
#> Adjustment method for p-values: bonferroni
#> # A tibble: 6 x 6
#> group1 group2 mean.difference p.value significance
#> <chr> <chr> <dbl> <dbl> <chr>
#> 1 carni herbi 0.542 1 ns
#> 2 carni insecti -0.0577 1 ns
#> 3 carni omni 0.0665 1 ns
#> 4 herbi insecti -0.600 1 ns
#> 5 herbi omni -0.476 0.979 ns
#> 6 insecti omni 0.124 1 ns
#> label
#> <chr>
#> 1 list(~italic(p)['adjusted']== 1.000 )
#> 2 list(~italic(p)['adjusted']== 1.000 )
#> 3 list(~italic(p)['adjusted']== 1.000 )
#> 4 list(~italic(p)['adjusted']== 1.000 )
#> 5 list(~italic(p)['adjusted']== 0.979 )
#> 6 list(~italic(p)['adjusted']== 1.000 )
# if `var.equal = FALSE`, then Games-Howell test will be run
pairwise_comparisons(
data = ggplot2::msleep,
x = vore,
y = brainwt,
type = "parametric",
var.equal = FALSE,
paired = FALSE,
p.adjust.method = "bonferroni"
)
#> Note: The parametric pairwise multiple comparisons test used-
#> Games-Howell test.
#> Adjustment method for p-values: bonferroni
#> # A tibble: 6 x 9
#> group1 group2 mean.difference se t.value df p.value significance
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 omni herbi 0.476 0.255 1.32 20.9 1 ns
#> 2 omni carni -0.066 0.061 0.774 21.1 1 ns
#> 3 omni insecti -0.124 0.057 1.55 17.2 1 ns
#> 4 herbi carni -0.542 0.25 1.54 19.4 1 ns
#> 5 herbi insecti -0.6 0.249 1.70 19.1 1 ns
#> 6 carni insecti -0.058 0.027 1.53 10.7 1 ns
#> label
#> <chr>
#> 1 list(~italic(p)['adjusted']== 1.000 )
#> 2 list(~italic(p)['adjusted']== 1.000 )
#> 3 list(~italic(p)['adjusted']== 1.000 )
#> 4 list(~italic(p)['adjusted']== 1.000 )
#> 5 list(~italic(p)['adjusted']== 1.000 )
#> 6 list(~italic(p)['adjusted']== 1.000 )
# non-parametric
pairwise_comparisons(
data = ggplot2::msleep,
x = vore,
y = brainwt,
type = "nonparametric",
paired = FALSE,
p.adjust.method = "none"
)
#> Note: The nonparametric pairwise multiple comparisons test used-
#> Dwass-Steel-Crichtlow-Fligner test.
#> Adjustment method for p-values: none
#> # A tibble: 6 x 6
#> group1 group2 W p.value significance
#> <chr> <chr> <dbl> <dbl> <chr>
#> 1 carni herbi -0.8 0.572 ns
#> 2 carni insecti -2.36 0.0956 ns
#> 3 carni omni -1.72 0.225 ns
#> 4 herbi insecti -2.40 0.0895 ns
#> 5 herbi omni -0.948 0.503 ns
#> 6 insecti omni 1.61 0.256 ns
#> label
#> <chr>
#> 1 list(~italic(p)['unadjusted']== 0.572 )
#> 2 list(~italic(p)['unadjusted']== 0.096 )
#> 3 list(~italic(p)['unadjusted']== 0.225 )
#> 4 list(~italic(p)['unadjusted']== 0.089 )
#> 5 list(~italic(p)['unadjusted']== 0.503 )
#> 6 list(~italic(p)['unadjusted']== 0.256 )
# robust
pairwise_comparisons(
data = ggplot2::msleep,
x = vore,
y = brainwt,
type = "robust",
paired = FALSE,
p.adjust.method = "fdr"
)
#> Note: The robust pairwise multiple comparisons test used-
#> Yuen's trimmed means comparisons test.
#> Adjustment method for p-values: fdr
#> # A tibble: 6 x 8
#> group1 group2 psihat conf.low conf.high p.value significance
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 insecti omni -0.0556 -0.184 0.0728 0.969 ns
#> 2 carni herbi -0.0530 -0.274 0.168 0.969 ns
#> 3 carni omni 0.00210 -0.151 0.155 0.969 ns
#> 4 herbi omni 0.0551 -0.173 0.283 0.969 ns
#> 5 carni insecti 0.0577 -0.0609 0.176 0.969 ns
#> 6 herbi insecti 0.111 -0.0983 0.320 0.969 ns
#> label
#> <chr>
#> 1 list(~italic(p)['adjusted']== 0.969 )
#> 2 list(~italic(p)['adjusted']== 0.969 )
#> 3 list(~italic(p)['adjusted']== 0.969 )
#> 4 list(~italic(p)['adjusted']== 0.969 )
#> 5 list(~italic(p)['adjusted']== 0.969 )
#> 6 list(~italic(p)['adjusted']== 0.969 )# for reproducibility
set.seed(123)
# parametric
pairwise_comparisons(
data = bugs_long,
x = condition,
y = desire,
type = "parametric",
paired = TRUE,
p.adjust.method = "BH"
)
#> Note: The parametric pairwise multiple comparisons test used-
#> Student's t-test.
#> Adjustment method for p-values: BH
#> # A tibble: 6 x 6
#> group1 group2 mean.difference p.value significance
#> <chr> <chr> <dbl> <dbl> <chr>
#> 1 HDHF HDLF -1.11 10.00e- 4 ***
#> 2 HDHF LDHF -0.474 7.07e- 2 ns
#> 3 HDHF LDLF -2.14 7.64e-12 ***
#> 4 HDLF LDHF 0.637 5.47e- 2 ns
#> 5 HDLF LDLF -1.03 1.39e- 3 **
#> 6 LDHF LDLF -1.66 6.67e- 9 ***
#> label
#> <chr>
#> 1 list(~italic(p)['adjusted']== 0.001 )
#> 2 list(~italic(p)['adjusted']== 0.071 )
#> 3 list(~italic(p)['adjusted']<= 0.001 )
#> 4 list(~italic(p)['adjusted']== 0.055 )
#> 5 list(~italic(p)['adjusted']== 0.001 )
#> 6 list(~italic(p)['adjusted']<= 0.001 )
# non-parametric
pairwise_comparisons(
data = bugs_long,
x = condition,
y = desire,
type = "nonparametric",
paired = TRUE,
p.adjust.method = "BY"
)
#> Note: The nonparametric pairwise multiple comparisons test used-
#> Durbin-Conover test.
#> Adjustment method for p-values: BY
#> # A tibble: 6 x 6
#> group1 group2 statistic p.value significance
#> <chr> <chr> <dbl> <dbl> <chr>
#> 1 HDHF HDLF 4.78 1.44e- 5 ***
#> 2 HDHF LDHF 2.44 4.47e- 2 *
#> 3 HDHF LDLF 8.01 5.45e-13 ***
#> 4 HDLF LDHF 2.34 4.96e- 2 *
#> 5 HDLF LDLF 3.23 5.05e- 3 **
#> 6 LDHF LDLF 5.57 4.64e- 7 ***
#> label
#> <chr>
#> 1 list(~italic(p)['adjusted']<= 0.001 )
#> 2 list(~italic(p)['adjusted']== 0.045 )
#> 3 list(~italic(p)['adjusted']<= 0.001 )
#> 4 list(~italic(p)['adjusted']== 0.050 )
#> 5 list(~italic(p)['adjusted']== 0.005 )
#> 6 list(~italic(p)['adjusted']<= 0.001 )
# robust
pairwise_comparisons(
data = bugs_long,
x = condition,
y = desire,
type = "robust",
paired = TRUE,
p.adjust.method = "hommel"
)
#> Note: The robust pairwise multiple comparisons test used-
#> Yuen's trimmed means comparisons test.
#> Adjustment method for p-values: hommel
#> # A tibble: 6 x 8
#> group1 group2 psihat conf.low conf.high p.value significance
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 HDLF LDHF -0.701 -1.71 0.303 6.20e- 2 ns
#> 2 HDHF LDHF 0.5 -0.188 1.19 6.20e- 2 ns
#> 3 HDLF LDLF 0.938 0.0694 1.81 1.36e- 2 *
#> 4 HDHF HDLF 1.16 0.318 2.00 1.49e- 3 **
#> 5 LDHF LDLF 1.54 0.810 2.27 1.16e- 6 ***
#> 6 HDHF LDLF 2.10 1.37 2.82 1.79e-10 ***
#> label
#> <chr>
#> 1 list(~italic(p)['adjusted']== 0.062 )
#> 2 list(~italic(p)['adjusted']== 0.062 )
#> 3 list(~italic(p)['adjusted']== 0.014 )
#> 4 list(~italic(p)['adjusted']== 0.001 )
#> 5 list(~italic(p)['adjusted']<= 0.001 )
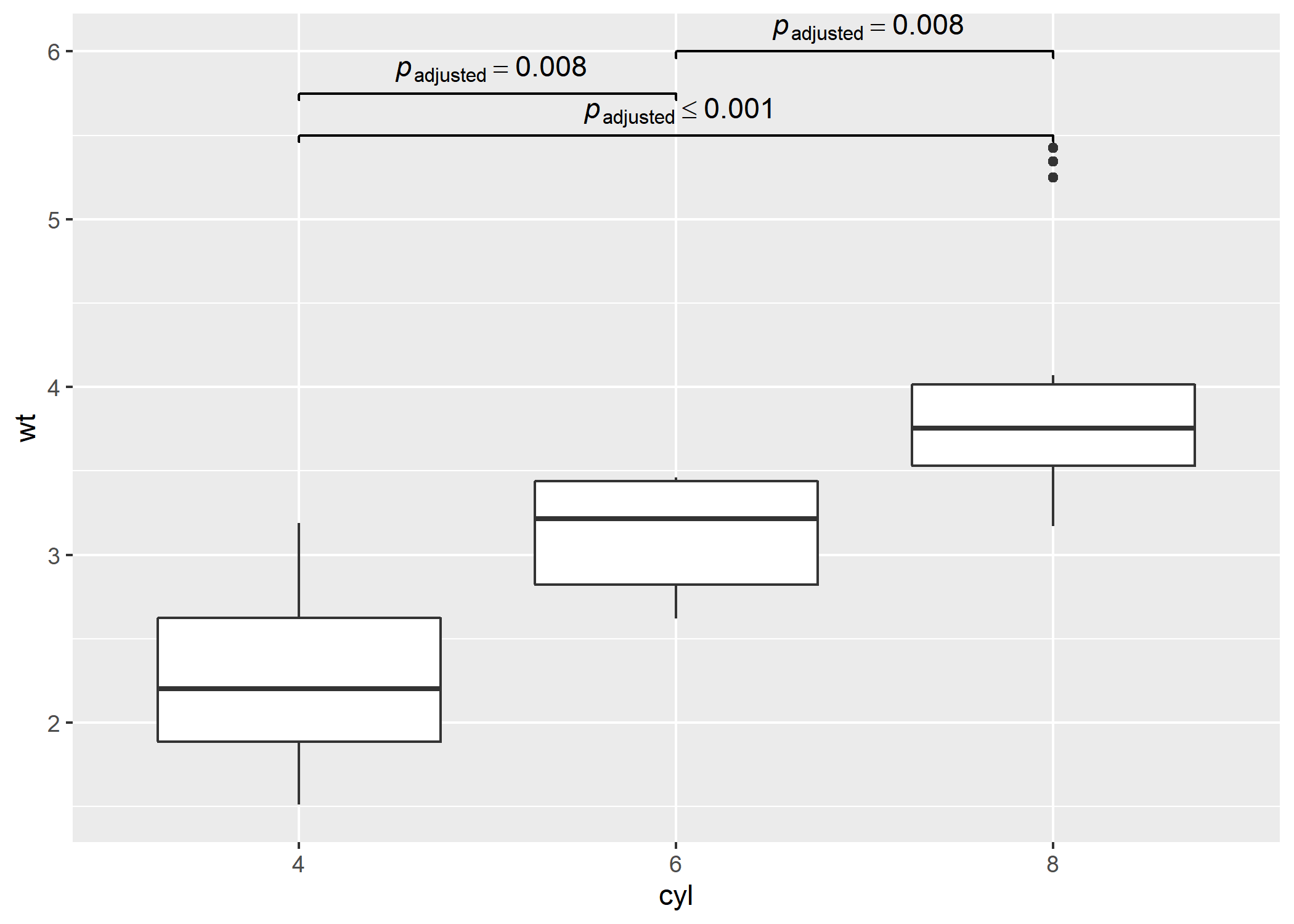
#> 6 list(~italic(p)['adjusted']<= 0.001 )# needed libraries
library(ggplot2)
library(pairwiseComparisons)
library(ggsignif)
# converting to factor
mtcars$cyl <- as.factor(mtcars$cyl)
# creating a basic plot
p <- ggplot(mtcars, aes(cyl, wt)) + geom_boxplot()
# using `pairwiseComparisons` package to create a dataframe with results
(df <-
pairwise_comparisons(mtcars, cyl, wt, messages = FALSE) %>%
dplyr::mutate(.data = ., groups = purrr::pmap(.l = list(group1, group2), .f = c)) %>%
dplyr::arrange(.data = ., group1))
#> # A tibble: 3 x 10
#> group1 group2 mean.difference se t.value df p.value significance
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 4 8 1.71 0.188 6.44 23.0 0 ***
#> 2 6 4 -0.831 0.154 3.81 16.0 0.008 **
#> 3 6 8 0.882 0.172 3.62 19.0 0.008 **
#> label groups
#> <chr> <list>
#> 1 list(~italic(p)['adjusted']<= 0.001 ) <chr [2]>
#> 2 list(~italic(p)['adjusted']== 0.008 ) <chr [2]>
#> 3 list(~italic(p)['adjusted']== 0.008 ) <chr [2]>
# using `geom_signif` to display results
p +
ggsignif::geom_signif(
comparisons = df$groups,
map_signif_level = TRUE,
tip_length = 0.01,
y_position = c(5.5, 5.75, 6),
annotations = df$label,
test = NULL,
na.rm = TRUE,
parse = TRUE
)As the code stands right now, here is the code coverage for all primary functions involved: https://codecov.io/gh/IndrajeetPatil/pairwiseComparisons/tree/master/R
I’m happy to receive bug reports, suggestions, questions, and (most of
all) contributions to fix problems and add features. I personally prefer
using the GitHub issues system over trying to reach out to me in other
ways (personal e-mail, Twitter, etc.). Pull Requests for contributions
are encouraged.
Here are some simple ways in which you can contribute (in the increasing order of commitment):
-
Read and correct any inconsistencies in the documentation
-
Raise issues about bugs or wanted features
-
Review code
-
Add new functionality (in the form of new plotting functions or helpers for preparing subtitles)
Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.