CPrior has functionalities to perform Bayesian statistics and running A/B and multivariate testing. CPrior includes several conjugate prior distributions and has been developed focusing on performance, implementing many closed-forms in terms of special functions to obtain fast and accurate results, avoiding Monte Carlo methods whenever possible.
Website: http://gnpalencia.org/cprior/
To install the current release of CPrior on Linux/Windows:
pip install cpriorFor different OS and/or custom installations, see http://gnpalencia.org/cprior/getting_started.html.
CPrior has been tested with CPython 3.5, 3.6 and 3.7. It requires:
- mpmath 1.0.0 or later. Website: http://mpmath.org/
- numpy 1.15.0 or later. Website: https://www.numpy.org/
- scipy 1.0.0 or later. Website: https://scipy.org/scipylib/
- pytest
- coverage
Run all unit tests
python setup.py testA Bayesian multivariate test with control and 3 variants. Data follows a Bernoulli distribution with distinct success probability.
- Generate control and variant models and build experiment. Select stopping rule and threshold (epsilon).
from scipy import stats
from cprior.models.bernoulli import BernoulliModel
from cprior.models.bernoulli import BernoulliMVTest
from cprior.experiment.base import Experiment
modelA = BernoulliModel(name="control", alpha=1, beta=1)
modelB = BernoulliModel(name="variation", alpha=1, beta=1)
modelC = BernoulliModel(name="variation", alpha=1, beta=1)
modelD = BernoulliModel(name="variation", alpha=1, beta=1)
mvtest = BernoulliMVTest({"A": modelA, "B": modelB, "C": modelC, "D": modelD})
experiment = Experiment(name="CTR", test=mvtest, stopping_rule="probability_vs_all",
epsilon=0.99, min_n_samples=1000, max_n_samples=None)- See experiment description
experiment.describe()=====================================================
Experiment: CTR
=====================================================
Bayesian model: bernoulli-beta
Number of variants: 4
Options:
stopping rule probability_vs_all
epsilon 0.99000
min_n_samples 1000
max_n_samples not set
Priors:
alpha beta
A 1 1
B 1 1
C 1 1
D 1 1
-------------------------------------------------- Generate or pass new data and update models until a clear winner is found. The stopping rule will be updated after a new update.
with experiment as e:
while not e.termination:
data_A = stats.bernoulli(p=0.0223).rvs(size=25)
data_B = stats.bernoulli(p=0.1128).rvs(size=15)
data_C = stats.bernoulli(p=0.0751).rvs(size=35)
data_D = stats.bernoulli(p=0.0280).rvs(size=15)
e.run_update(**{"A": data_A, "B": data_B, "C": data_C, "D": data_D})
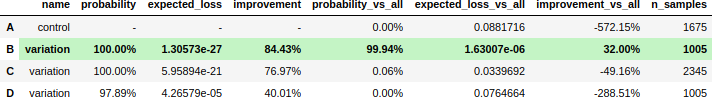
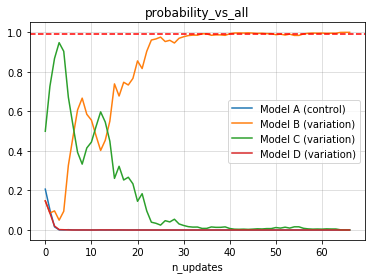
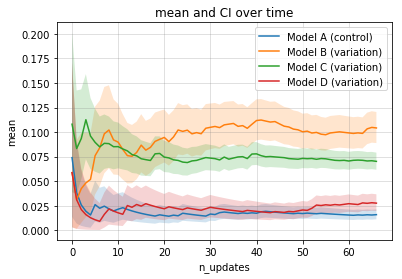
print(e.termination, e.status)True winner B- Reporting: experiment summary
experiment.summary()- Reporting: visualize stopping rule metric over time (updates)
experiment.plot_metric()- Reporting: visualize statistics over time (updates)
experiment.plot_stats()A Bayesian A/B test with data following a Bernoulli distribution with two distinct success probability. This example is a simple use case for CRO (conversion rate) or CTR (click-through rate) testing.
from scipy import stats
from cprior.models import BernoulliModel
from cprior.models import BernoulliABTest
modelA = BernoulliModel()
modelB = BernoulliModel()
test = BernoulliABTest(modelA=modelA, modelB=modelB)
data_A = stats.bernoulli(p=0.10).rvs(size=1500, random_state=42)
data_B = stats.bernoulli(p=0.11).rvs(size=1600, random_state=42)
test.update_A(data_A)
test.update_B(data_B)
# Compute P[A > B] and P[B > A]
print("P[A > B] = {:.4f}".format(test.probability(variant="A")))
print("P[B > A] = {:.4f}".format(test.probability(variant="B")))
# Compute posterior expected loss given a variant
print("E[max(B - A, 0)] = {:.4f}".format(test.expected_loss(variant="A")))
print("E[max(A - B, 0)] = {:.4f}".format(test.expected_loss(variant="B")))The output should be the following:
P[A > B] = 0.1024 P[B > A] = 0.8976 E[max(B - A, 0)] = 0.0147 E[max(A - B, 0)] = 0.0005