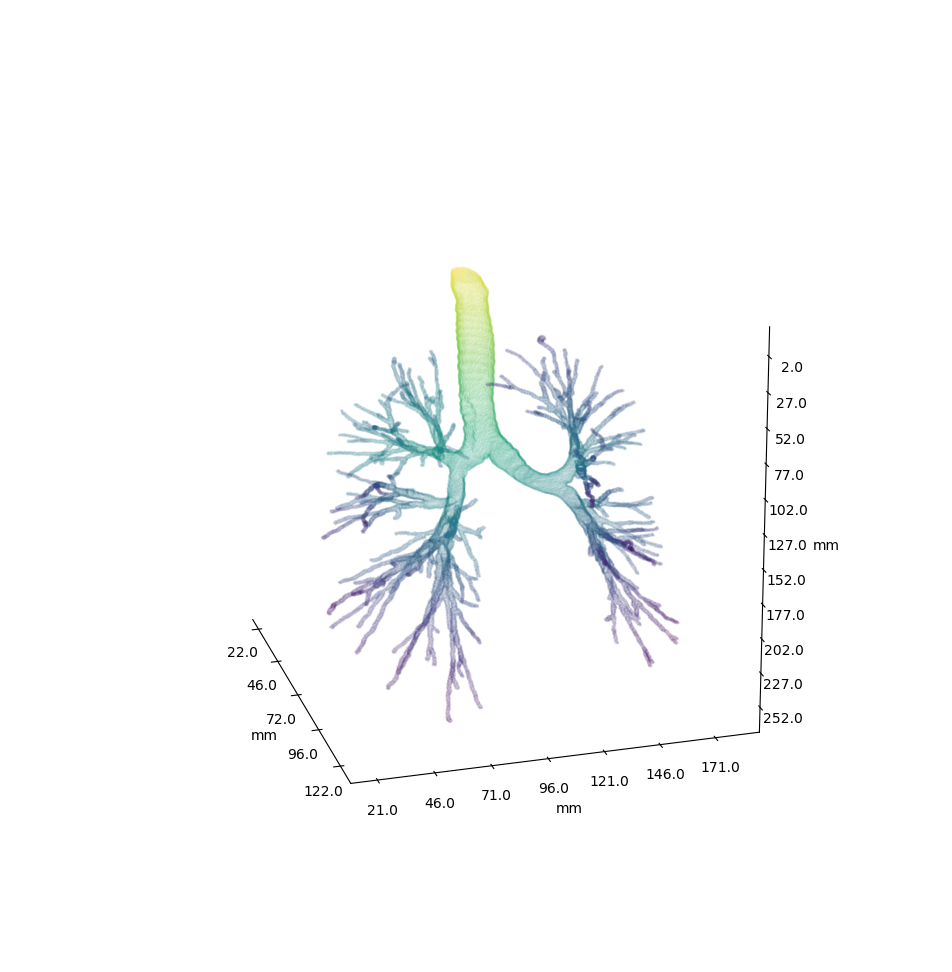
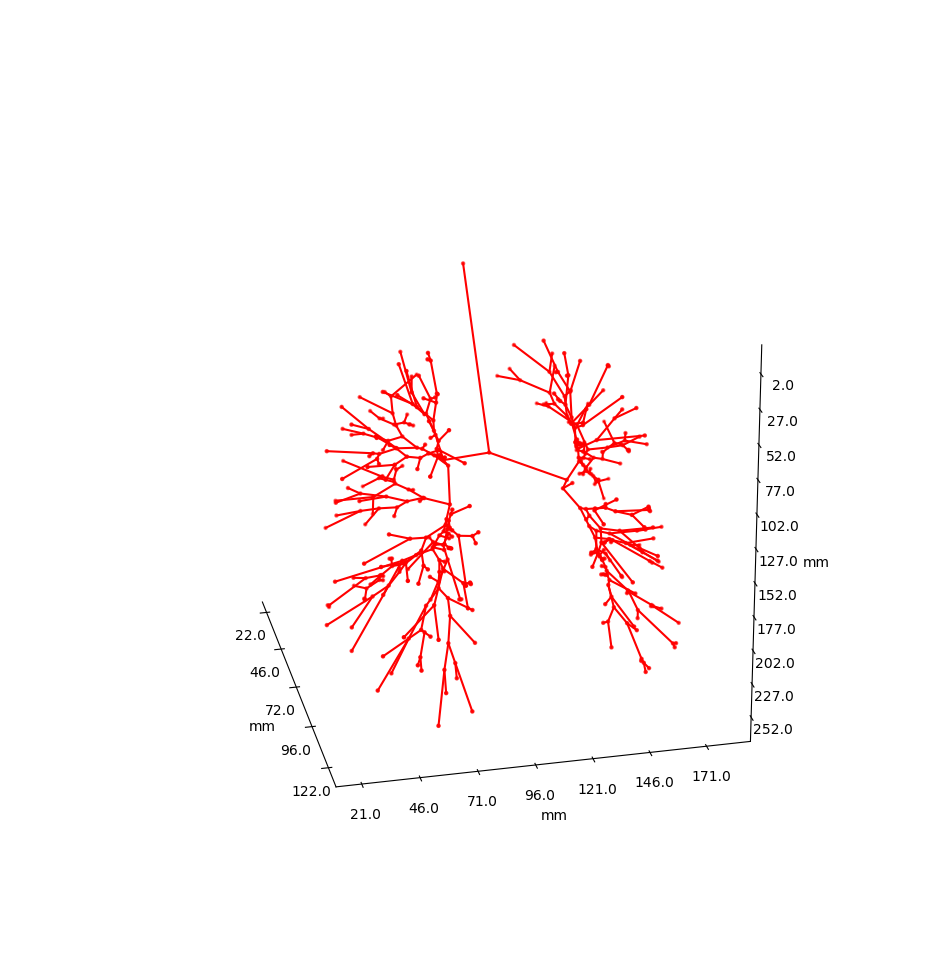
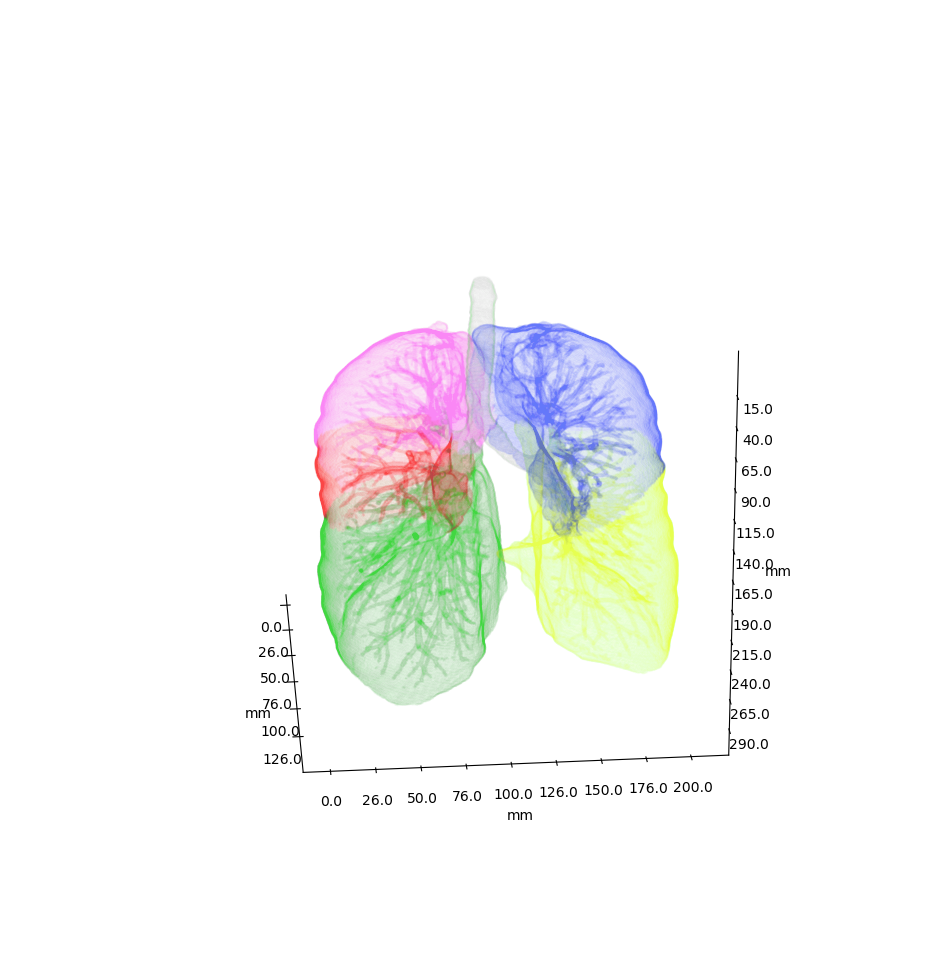
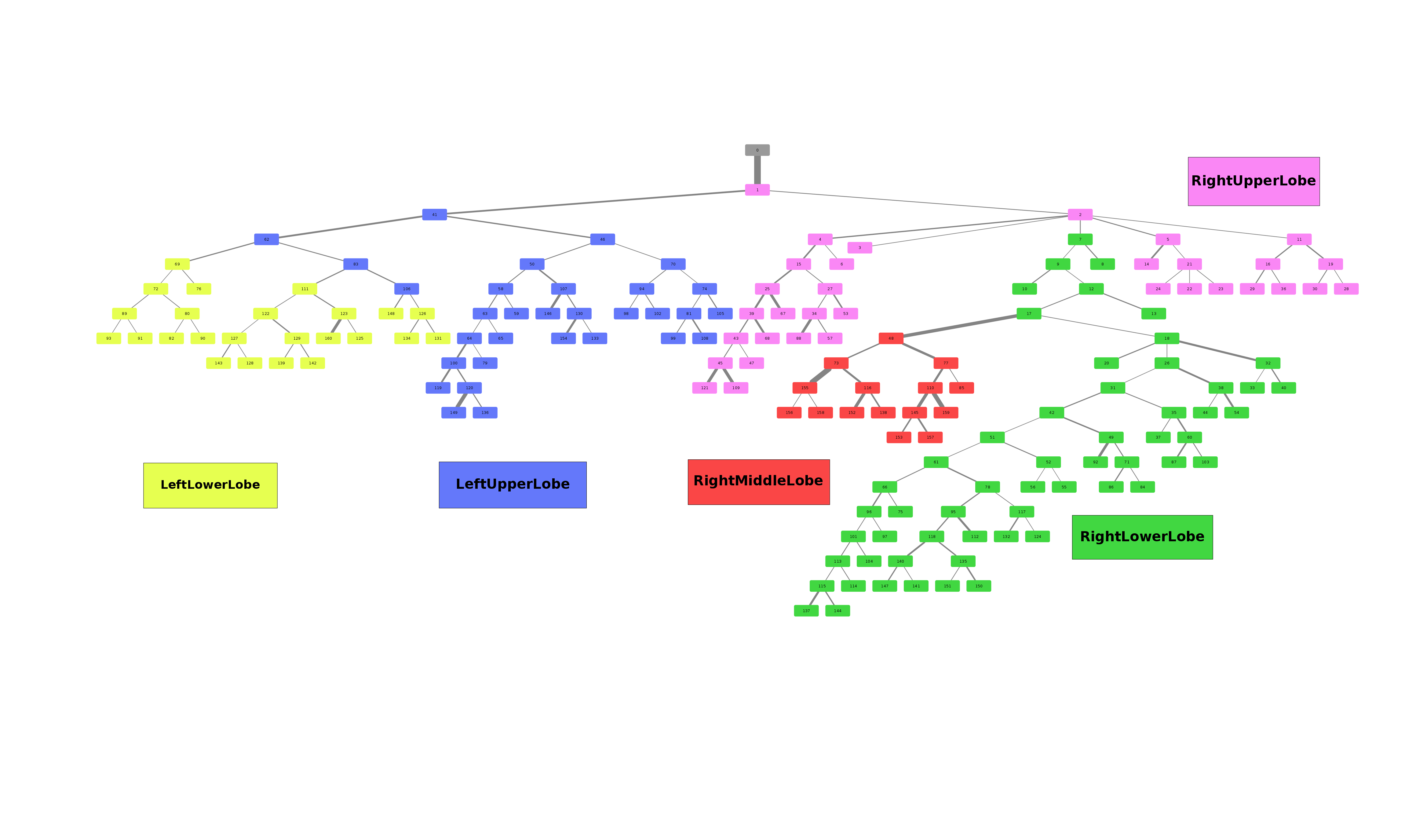
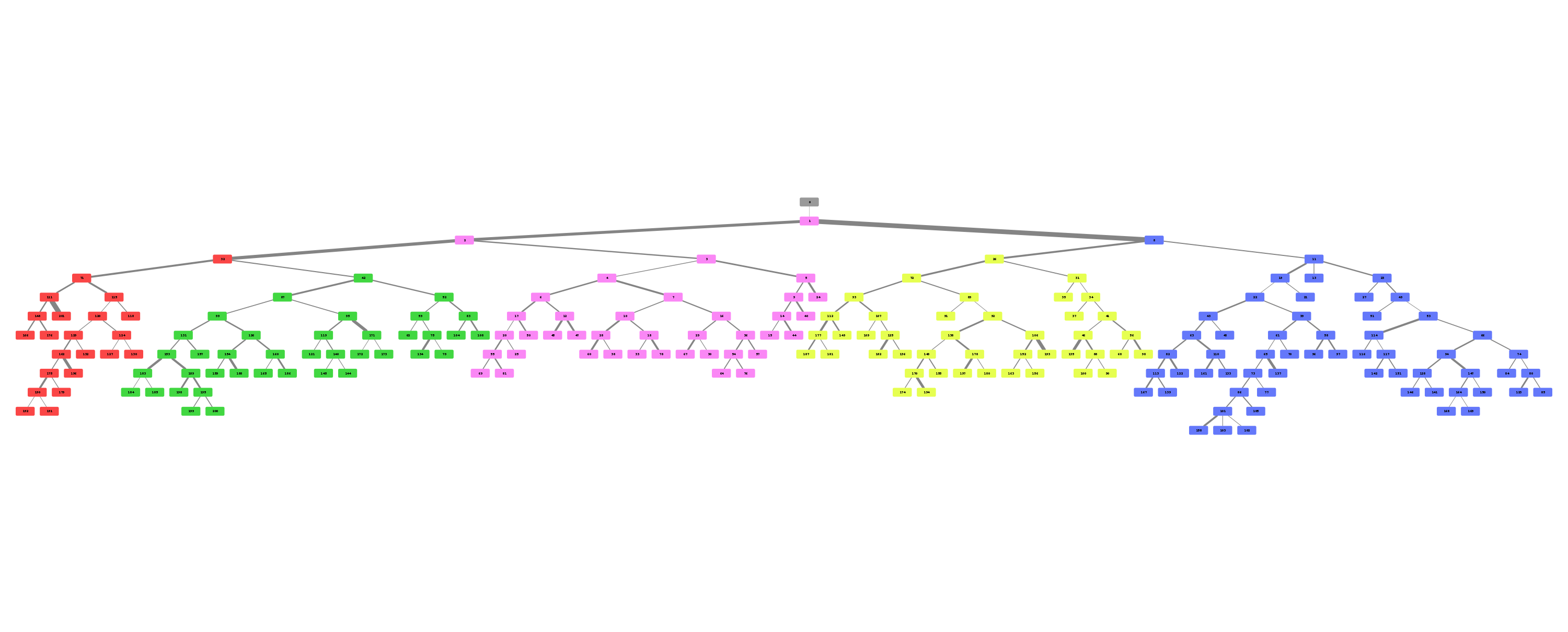
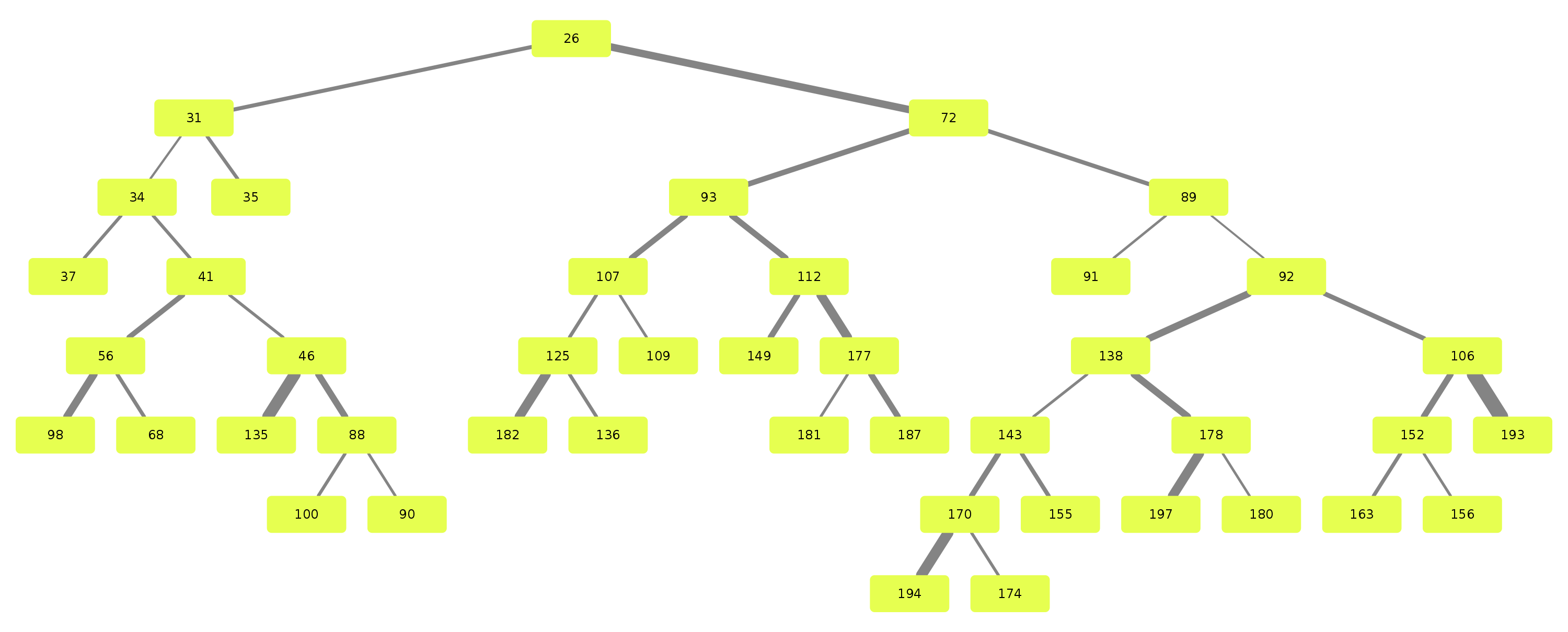
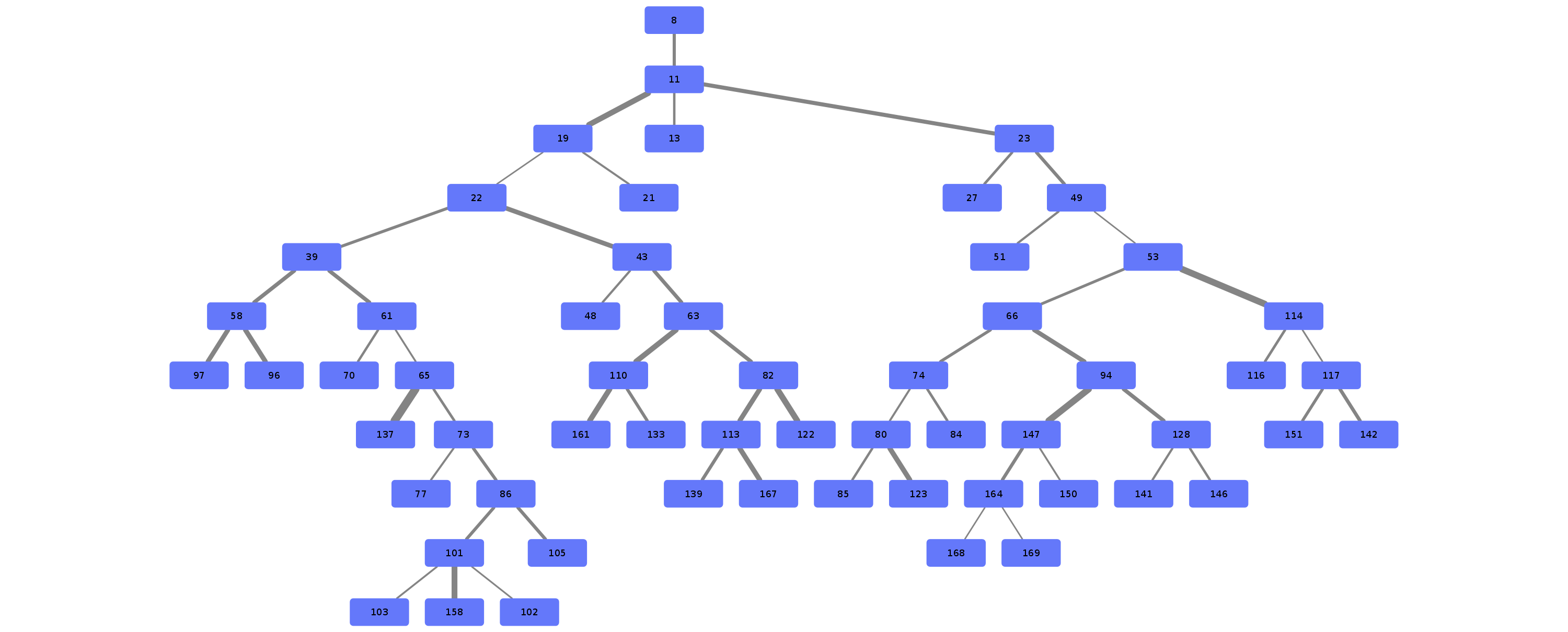
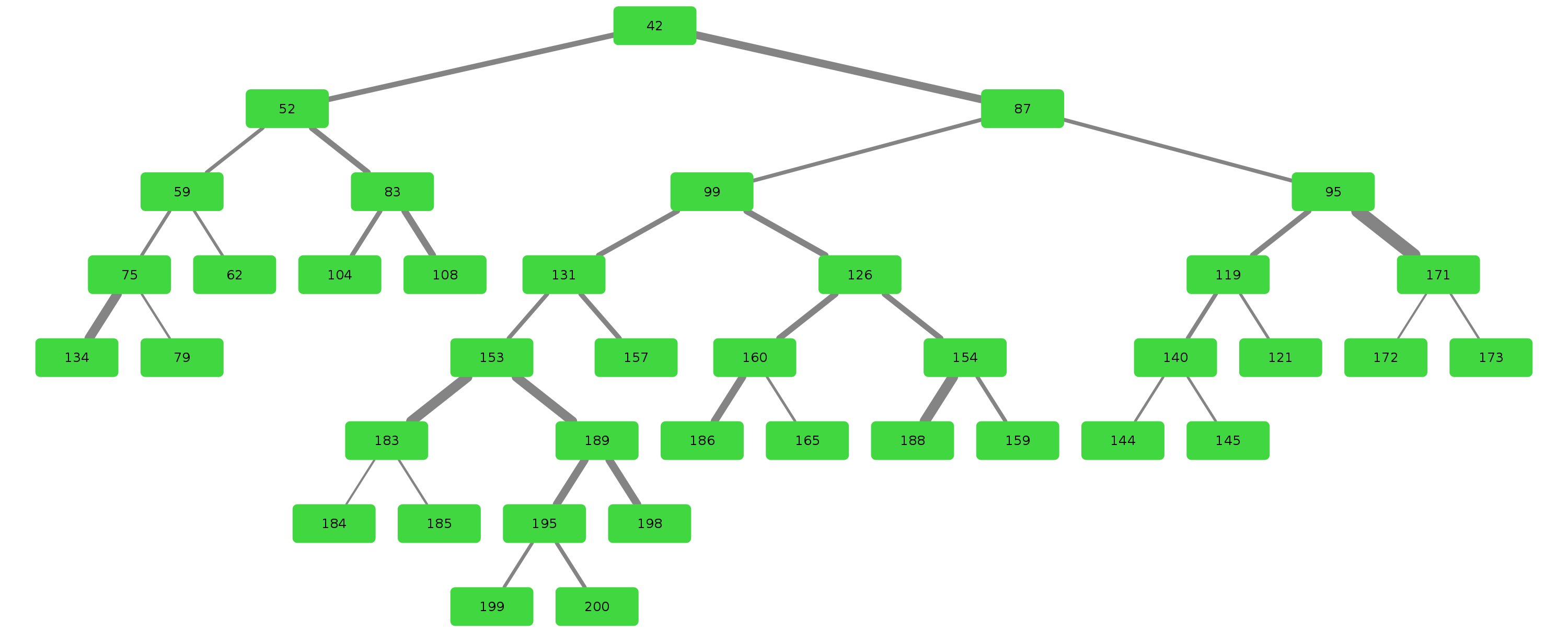
Airway is a project which analyses the anatomy of the lung bronchus by viewing it as a tree and looking for unusual structures. Specifically the upper left lobe is of interest, where there is a rather unusual three-way split after the lingula.
According to Dr. Rolf Oerter this has not yet been documented and is of great interest to surgeons.
This project is WIP and currently very actively in development!
We use a pipeline based approach to calculate the raw data. With the help of
airway-pipeline.py you are able to calculate each step (called stages) separately or
all at once.
Everything happens in a directory of your choice. Prerequisites are the raw data and the filesystem structure of the raw data.
Here is how the data has to be defined. Note that raw_data should be formatted
to raw_airway by you as we do not know your initial data structure.
We do provide a script which handled our case, you can find it in
scripts/separate-bronchus-files.sh.
DATADIR
├── raw_data <- This is the entirely unformatted raw data
│ └── Ct_Thorax_Standard_Mit_Km - 3123156
│ ├── DATA
│ │ ├── Data.txt <- This contained the paths for finding the various bronchus and lobes
│ │ └── 3123156 <- Example Patient ID
│ │ └── ... <- Some folder structure with the data
├── raw_airway <- Formatted data which will be used as input for stage-01
│ └── 3123156
│ ├── Artery
│ ├── Bronchus
│ ├── Koerperstamm
│ ├── LeftLowerLobe
│ ├── LeftUpperLobe
│ ├── RightLowerLobe
│ ├── RightMiddleLobe
│ ├── RightUpperLobe
│ └── Vein
├── stage-01 <- Each stage now has the same basic format
│ ├── 3123156
│ └── ...
├── stage-02
│ ├── 3123156
│ └── ...
...
After you have created the raw_airway folder structure, you can use the airway-pipeline.py script to create various stages.
To do this rename example_defaults.yaml to defaults.yaml and change the path in the file to where you have put the data.
You may ignore the other parameters for now.
For every calculated stage airway-pipeline.py creates a new directory (stage-xx) and subdirectories for each patient (e.g. 3123156).
Let us calculate stage 1:
./airway-pipeline.py 1
The airway pipeline checks if the stage already exists, if you need to overwrite
a stage you need to add the -f/--force flag.
Here are some other possible ways to define stages:
./airway-pipeline.py 1 3-4 6+ - will create stages 1, 3, 4, 6 and all the ones above 6
./airway-pipeline.py tree vis - will create stages which are in the groups tree and vis
./airway-pipeline.py all - will create all stages
By default four patients will be calculated in parallel (4 workers are used). If you have more CPU threads, simply increase the number of workers:
./airway-pipeline.py 1 2 3 -w 32 or change the default in the config file (defaults.yaml).
You can type ./airway-pipeline --stages for this description as well:
TODO: out of date
Stages 1 through 9 are responsible for generation of the bronchus tree splits.
Stage 10 through 19 are responsible for tree analysis.
Stages 20 through 29 are responsible for visualisation.
Airway originated as an observation by Dr. Rolf Oerter at the University of Rostock that certain structures in the lungs bronchus he has seen while operating have not been documented. The first steps of the project were made as a student project at the University of Rostock at the Department of Systems Biology organised by Mariam Nassar.
It consisted of this Team:
- Martin Steinbach
- Brutenis Gliwa
- Lukas Großehagenbrock
- Jonas Moesicke
- Joris Thiele
After this, the project is being continued by me (Brutenis) as my bachelor thesis. Thanks to Mariam Nassar, Gundram Leifert and Prof. Olaf Wolkenhauer for supervision during this time. And thanks to Planet AI for letting me write my thesis at their office.