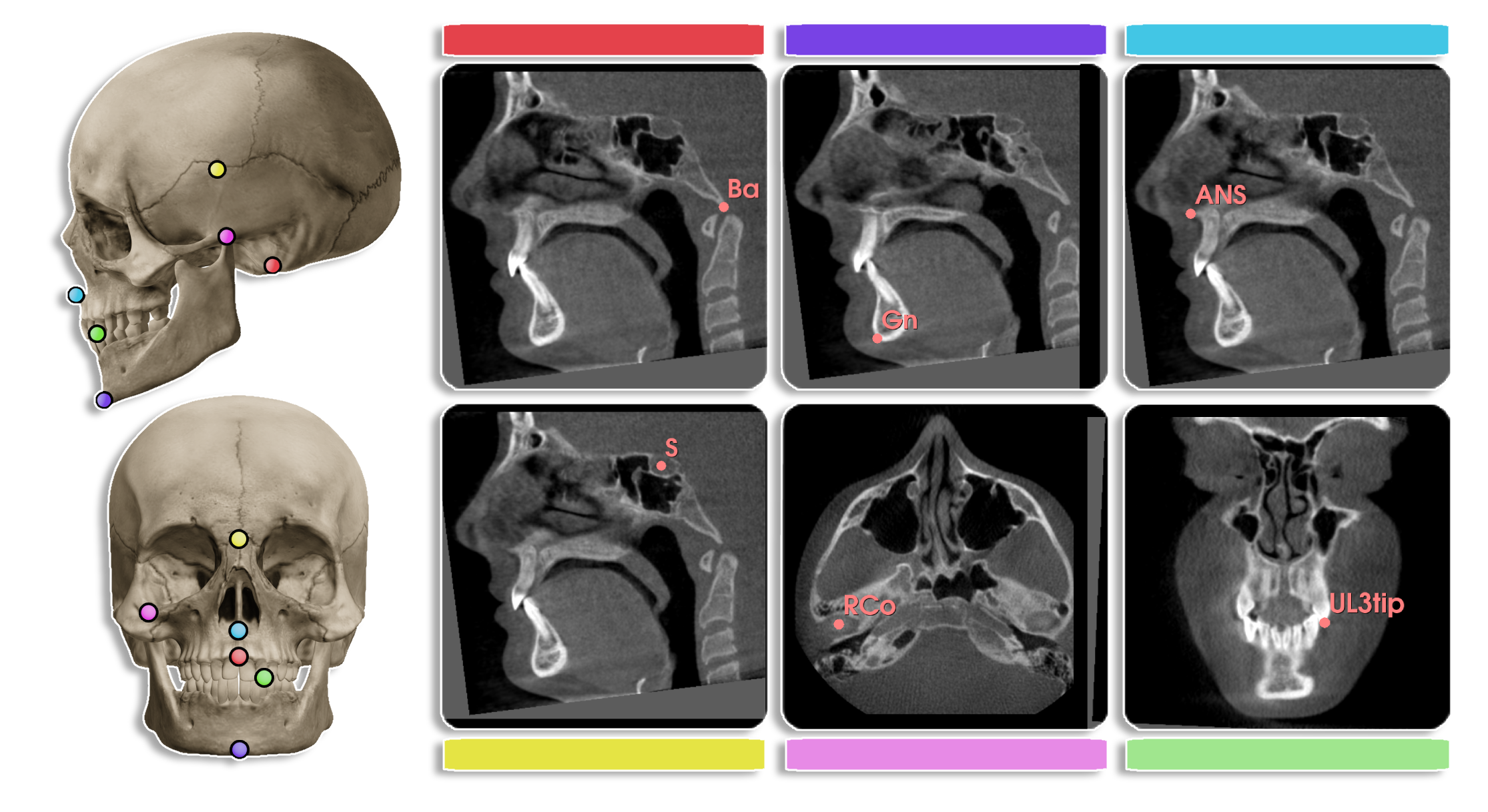
Cephalometric analysis is a diagnostic tool used in orthodontics to measure the size and spatial relationships of the teeth, jaws, and cranium. It is used to diagnose and plan treatment for patients with dentofacial abnormalities.
The central problem in cephalometric analysis is to identify the landmarks in the cone-beam computed tomography (CBCT) volume. Due to the large size of the CBCT volume, manual identification of landmarks is time-consuming and error-prone. Automated landmark detection can help in reducing the time and effort required for cephalometric analysis, such as analyzing the symmetry of the face, measuring the size of the jaw, and diagnosing the position of the teeth.
The original paper (Influence of growth structures and fixed appliances on automated cephalometric landmark recognition with a customized convolutional neural network) aim to detection the key landmarks in the 2D cephalometric images. And extend the work to 3D CBCT volumes is a natural next step [1].
-
PDDCA dataset is a Public Domain Database for Computational Anatomy. It contains 48 patient CT volumes, with manual segmentation and bony landmarks. The size of the images is 512 × 512 x 384 voxels, with a voxel size of 0.9766 × 0.9766 x 1.25 mm3.
-
CQ500 Dataset is a dataset of 491 head CT scans, with label of abnormalities. This dataset can be used for pretraining models for landmark detection.
-
Almost every paper contains datasets that are private and not publicly available. The size of the datasets varies from 10 to 200 CBCT volumes.
The overall pipeline for landmark detection in CBCT volumes is as follows [4,5,6,7]:
- Coarse grained detection of the region of interest (ROI) containing the key landmarks.
- Fine identification of the key landmarks within the ROI.
The coarse grained detection of the ROI can be done using a 3D U-Net model, resulting in a heatmap or bouding box around the key landmarks. The fine model takes the ROI as input and outputs the key landmarks.
-
OrthoLab from U of Michigan provide segmentation and landmark detection models for 2D cephalometric images. they trained a UNet-Tranformer model on 618 de-identified CBCT volumes data.

They also provide 3D keypoint detection model for CBCT volumes (MIDL 2022).
-
What are clinical analysis that can be done using cephalometric analysis? Can we automate these analysis?
-
A technical question could be improving the efficiency of the landmark detection model.
-
Why is it limited work on using registration-based methods for landmark detection in CBCT volumes? It seems that the overral structure of the skull is similar across patients, so registration-based methods require less data.
-
What are the plan from the authors with the problem, are they going to publish a new dataset, implement a new software, or improve the model?
-
Current generalize models are capable of segmenting the skull, can we use those model to minimize the ROI for the landmark detection model?
-
Private datasets prevent analysis of the generalization of the model across ethnicities, how can we address this issue?
-
Chien, P. C., E. T. Parks, Francisco Eraso, J. K. Hartsfield, W. E. Roberts, and S. Ofner. "Comparison of reliability in anatomical landmark identification using two-dimensional digital cephalometrics and three-dimensional cone beam computed tomography in vivo." Dentomaxillofacial Radiology. 2009.
-
Montúfar, Jesús, Marcelo Romero, and Rogelio J. Scougall-Vilchis. "Hybrid approach for automatic cephalometric landmark annotation on cone-beam computed tomography volumes.". American Journal of Orthodontics and Dentofacial Orthopedics.
-
Gillot, Maxime, Baptiste Baquero, Celia Le, Romain Deleat-Besson, Jonas Bianchi, Antonio Ruellas, Marcela Gurgel et al. "Automatic multi-anatomical skull structure segmentation of cone-beam computed tomography scans using 3D UNETR." Plos one. 2022.
-
Gillot, Maxime Paul Guy, Baptiste Antoine Baquero, Juan Carlos Prieto, and Lucia Cevidanes. "Automatic Landmark Identification in 3D Cone-Beam Computed Tomography scans." MIDL. 2021.
-
Chen R, Ma Y, Chen N et al "Structure-aware long shortterm memory network for 3D cephalometric landmark detection." IEEE Trans Med Imaging. 2022.
-
Lang Y, Lian C, Xiao D et al Localization of craniomaxillofacial landmarks on CBCT images using 3D mask R-CNN and local dependency learning. IEEE Trans Med Imaging. 2022.
-
Chen X, Lian C, Deng HH et al Fast and accurate craniomaxillofacial landmark detection via 3D Faster R-CNN. IEEE Trans Med Imaging. 2021.