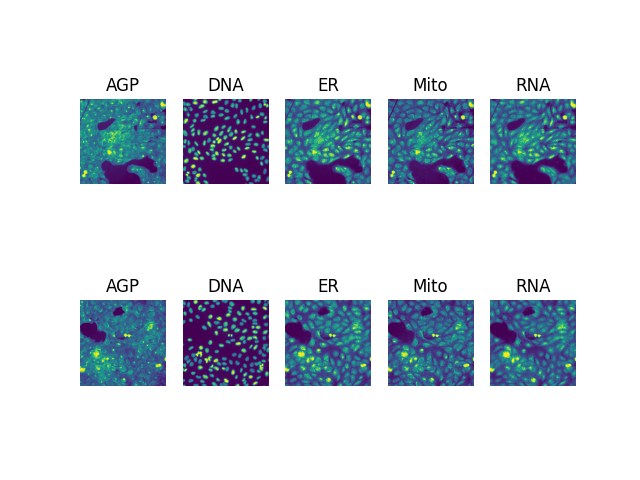
This is a repository aimed at downloading the JUMP Cell Painting dataset, which is a large dataset of molecule/image pairs obtained using High Content Screening, and can be used to predict molecular properties from images, and vice versa. The data can then be used to train downstream deep learning models.
The data follows the Cell Painting protocol, a common effort between multiple labs that contains more than 116k tested compound for a equivalent of more that 3M images.
This repository was created once the code was cleaned, the entire Git history is still available on another private repo.
Slides presenting the scenarios: Slides
We used the JUMP Cell Painting datasets (Chandrasekaran et al., 2023), available from the Cell Painting Gallery on the Registry of Open Data on AWS https://registry.opendata.aws/cellpainting-gallery/.
Chandrasekaran, S. N., Ackerman, J., Alix, E., Ando, D. M., Arevalo, J., Bennion, M., ... & Carpenter, A. E. (2023). JUMP Cell Painting dataset: morphological impact of 136,000 chemical and genetic perturbations. bioRxiv, 2023-03: 2023-03. doi:10.1101/2023.03.23.534023
jump
├── images
│ ├── source_1
│ │ ├── Batch1_20221004
│ │ │ ├── UL001641
│ │ │ │ ├── source_1__UL001641__A02__1__AGP.png
│ │ │ │ ├── source_1__UL001641__A02__1__DNA.png
│ │ │ │ ├── source_1__UL001641__A02__1__ER.png
│ │ │ │ ├── source_1__UL001641__A02__1__Mito.png
│ │ │ │ ├── source_1__UL001641__A02__1__RNA.png
│ │ │ │ ├── source_1__UL001641__A02__2__AGP.png
│ │ │ │ ├── source_1__UL001641__A02__2__DNA.png
│ │ │ │ ├── source_1__UL001641__A02__2__ER.png
│ │ │ │ ├── source_1__UL001641__A02__2__Mito.png
│ │ │ │ ├── source_1__UL001641__A02__2__RNA.png
│ │ │ │ └── ...
│ │ │ └── ...
│ │ └── ...
│ ├── source_13
│ ├── source_4
│ └── source_9
├── jobs
│ ├── ids
│ ├── submission.csv
│ ├── submissions_left.csv
│ └── test_submission.csv
├── load_data
│ ├── check
│ ├── final
│ ├── load_data_with_metadata
│ ├── load_data_with_samples
│ ├── total_illum.csv.gz
│ └── total_load_data.csv.gz
├── metadata
│ ├── complete_metadata.csv
│ ├── compound.csv
│ ├── compound.csv.gz
│ ├── crispr.csv
│ ├── crispr.csv.gz
│ ├── JUMP-Target-1_compound_metadata.tsv
│ ├── JUMP-Target-1_compound_platemap.tsv
│ ├── JUMP-Target-1_crispr_metadata.tsv
│ ├── JUMP-Target-1_crispr_platemap.tsv
│ ├── JUMP-Target-1_orf_metadata.tsv
│ ├── JUMP-Target-1_orf_platemap.tsv
│ ├── JUMP-Target-2_compound_metadata.tsv
│ ├── JUMP-Target-2_compound_platemap.tsv
│ ├── microscope_config.csv
│ ├── microscope_filter.csv
│ ├── orf.csv
│ ├── orf.csv.gz
│ ├── plate.csv
│ ├── plate.csv.gz
│ ├── README.md
│ ├── resolution.csv
│ ├── well.csv
│ └── well.csv.gz
└── README.mdThe most important files and folders are:
images: a folder containing images from the dataset, organized by source, batch and platemetadata: a folder containing metadata about the perturbationsmetadata/complete_metadata.csv: a file containing all the well-level metadata for the dataset. It is the merge of the following dataframes:metadata/plate.csv: a file containing a list of all plates in the datasetmetadata/well.csv: a file containing a list of all wells in the dataset, with their corresponding plate and perturbation IDmetadata/compound.csv: a file containing a list of all compound perturbationsmetadata/crispr.csv: a file containing a list of all CRISPR perturbationsmetadata/orf.csv: a file containing a list of all ORF perturbations
load_data: a folder containing the path to the images both locally and in the s3 bucketload_data/load_data_with_samples: a parquet dataframe (partitionned by source) containing the path to the images in the s3 bucket, as well as the filter column, indicating whether the image is kept or not in the local datasetload_data/final: a parquet dataframe (partitionned by plate) containing the path to the images locally. It only contains the images that are kept in the local dataset and doesn't include any metadata
jobs: a folder containing the jobs used with HTCondor
| Metadata_Source | Metadata_Batch | Metadata_Plate | Metadata_Well | Metadata_Site | FileName_OrigAGP | FileName_OrigDNA | FileName_OrigER | FileName_OrigMito | FileName_OrigRNA | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | source_9 | 20211103-Run16 | GR00004416 | A01 | 1 | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__1__AGP.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__1__DNA.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__1__ER.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__1__Mito.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__1__RNA.png |
| 1 | source_9 | 20211103-Run16 | GR00004416 | A01 | 3 | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__3__AGP.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__3__DNA.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__3__ER.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__3__Mito.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__3__RNA.png |
| 2 | source_9 | 20211103-Run16 | GR00004416 | A01 | 4 | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__4__AGP.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__4__DNA.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__4__ER.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__4__Mito.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A01__4__RNA.png |
| 3 | source_9 | 20211103-Run16 | GR00004416 | A02 | 1 | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__1__AGP.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__1__DNA.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__1__ER.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__1__Mito.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__1__RNA.png |
| 4 | source_9 | 20211103-Run16 | GR00004416 | A02 | 2 | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__2__AGP.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__2__DNA.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__2__ER.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__2__Mito.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__2__RNA.png |
| 5 | source_9 | 20211103-Run16 | GR00004416 | A02 | 3 | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__3__AGP.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__3__DNA.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__3__ER.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__3__Mito.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__3__RNA.png |
| 6 | source_9 | 20211103-Run16 | GR00004416 | A02 | 4 | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__4__AGP.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__4__DNA.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__4__ER.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__4__Mito.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A02__4__RNA.png |
| 7 | source_9 | 20211103-Run16 | GR00004416 | A03 | 1 | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__1__AGP.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__1__DNA.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__1__ER.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__1__Mito.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__1__RNA.png |
| 8 | source_9 | 20211103-Run16 | GR00004416 | A03 | 2 | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__2__AGP.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__2__DNA.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__2__ER.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__2__Mito.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__2__RNA.png |
| 9 | source_9 | 20211103-Run16 | GR00004416 | A03 | 3 | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__3__AGP.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__3__DNA.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__3__ER.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__3__Mito.png | /projects/cpjump1/jump/images/source_9/20211103-Run16/GR00004416/source_9__GR00004416__A03__3__RNA.png |
| Metadata_Source | Metadata_Batch | Metadata_Plate | Metadata_PlateType | Metadata_Well | Metadata_JCP2022 | Metadata_InChIKey | Metadata_InChI | Metadata_broad_sample | Metadata_Name | Metadata_Vector | Metadata_Transcript | Metadata_Symbol_x | Metadata_NCBI_Gene_ID_x | Metadata_Taxon_ID | Metadata_Gene_Description | Metadata_Prot_Match | Metadata_Insert_Length | Metadata_pert_type | Metadata_NCBI_Gene_ID_y | Metadata_Symbol_y | Metadata_Microscope_Name | Metadata_Widefield_vs_Confocal | Metadata_Excitation_Type | Metadata_Objective_NA | Metadata_N_Brightfield_Planes_Min | Metadata_N_Brightfield_Planes_Max | Metadata_Distance_Between_Z_Microns | Metadata_Sites_Per_Well | Metadata_Filter_Configuration | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | source_1 | Batch1_20221004 | UL000109 | COMPOUND_EMPTY | A02 | JCP2022_033924 | IAZDPXIOMUYVGZ-UHFFFAOYSA-N | InChI=1S/C2H6OS/c1-4(2)3/h1-2H3 | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | Opera Phenix | Widefield | Laser | 1 | 1 | 1 | nan | 4 | H |
| 1 | source_1 | Batch1_20221004 | UL000109 | COMPOUND_EMPTY | A03 | JCP2022_085227 | SRVFFFJZQVENJC-UHFFFAOYSA-N | InChI=1S/C17H30N2O5/c1-6-23-17(22)14-13(24-14)16(21)19-12(9-11(4)5)15(20)18-8-7-10(2)3/h10-14H,6-9H2,1-5H3,(H,18,20)(H,19,21) | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | Opera Phenix | Widefield | Laser | 1 | 1 | 1 | nan | 4 | H |
| 2 | source_1 | Batch1_20221004 | UL000109 | COMPOUND_EMPTY | A04 | JCP2022_033924 | IAZDPXIOMUYVGZ-UHFFFAOYSA-N | InChI=1S/C2H6OS/c1-4(2)3/h1-2H3 | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | Opera Phenix | Widefield | Laser | 1 | 1 | 1 | nan | 4 | H |
| 3 | source_1 | Batch1_20221004 | UL000109 | COMPOUND_EMPTY | A05 | JCP2022_033924 | IAZDPXIOMUYVGZ-UHFFFAOYSA-N | InChI=1S/C2H6OS/c1-4(2)3/h1-2H3 | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | Opera Phenix | Widefield | Laser | 1 | 1 | 1 | nan | 4 | H |
| 4 | source_1 | Batch1_20221004 | UL000109 | COMPOUND_EMPTY | A06 | JCP2022_033924 | IAZDPXIOMUYVGZ-UHFFFAOYSA-N | InChI=1S/C2H6OS/c1-4(2)3/h1-2H3 | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | Opera Phenix | Widefield | Laser | 1 | 1 | 1 | nan | 4 | H |
| 5 | source_1 | Batch1_20221004 | UL000109 | COMPOUND_EMPTY | A07 | JCP2022_033924 | IAZDPXIOMUYVGZ-UHFFFAOYSA-N | InChI=1S/C2H6OS/c1-4(2)3/h1-2H3 | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | Opera Phenix | Widefield | Laser | 1 | 1 | 1 | nan | 4 | H |
| 6 | source_1 | Batch1_20221004 | UL000109 | COMPOUND_EMPTY | A08 | JCP2022_033924 | IAZDPXIOMUYVGZ-UHFFFAOYSA-N | InChI=1S/C2H6OS/c1-4(2)3/h1-2H3 | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | Opera Phenix | Widefield | Laser | 1 | 1 | 1 | nan | 4 | H |
| 7 | source_1 | Batch1_20221004 | UL000109 | COMPOUND_EMPTY | A09 | JCP2022_033924 | IAZDPXIOMUYVGZ-UHFFFAOYSA-N | InChI=1S/C2H6OS/c1-4(2)3/h1-2H3 | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | Opera Phenix | Widefield | Laser | 1 | 1 | 1 | nan | 4 | H |
| 8 | source_1 | Batch1_20221004 | UL000109 | COMPOUND_EMPTY | A10 | JCP2022_033924 | IAZDPXIOMUYVGZ-UHFFFAOYSA-N | InChI=1S/C2H6OS/c1-4(2)3/h1-2H3 | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | Opera Phenix | Widefield | Laser | 1 | 1 | 1 | nan | 4 | H |
| 9 | source_1 | Batch1_20221004 | UL000109 | COMPOUND_EMPTY | A11 | JCP2022_033924 | IAZDPXIOMUYVGZ-UHFFFAOYSA-N | InChI=1S/C2H6OS/c1-4(2)3/h1-2H3 | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | Opera Phenix | Widefield | Laser | 1 | 1 | 1 | nan | 4 | H |
| broad_sample | InChIKey | pert_iname | pubchem_cid | target | pert_type | control_type | smiles | |
|---|---|---|---|---|---|---|---|---|
| 0 | BRD-A86665761-001-01-1 | TZDUHAJSIBHXDL-UHFFFAOYSA-N | gabapentin-enacarbil | 9883933 | CACNB4 | trt | nan | CC(C)C(=O)OC(C)OC(=O)NCC1(CC(O)=O)CCCCC1 |
| 1 | BRD-A22032524-074-09-9 | HTIQEAQVCYTUBX-UHFFFAOYSA-N | amlodipine | 2162 | CACNA2D3 | trt | nan | CCOC(=O)C1=C(COCCN)NC(C)=C(C1c1ccccc1Cl)C(=O)OC |
| 2 | BRD-A01078468-001-14-8 | PBBGSZCBWVPOOL-UHFFFAOYSA-N | hexestrol | 3606 | AKR1C1 | trt | nan | CCC(C(CC)c1ccc(O)cc1)c1ccc(O)cc1 |
| 3 | BRD-K48278478-001-01-2 | LOUPRKONTZGTKE-AFHBHXEDSA-N | quinine | 94175 | KCNN4 | trt | nan | COc1ccc2nccc(C@@H[C@H]3C[C@@H]4CC[N@]3C[C@@H]4C=C)c2c1 |
| 4 | BRD-K36574127-001-01-3 | NYNZQNWKBKUAII-KBXCAEBGSA-N | LOXO-101 | 46188928 | NTRK1 | trt | nan | O[C@H]1CCN(C1)C(=O)Nc1cnn2ccc(nc12)N1CCC[C@@H]1c1cc(F)ccc1F |
| 5 | BRD-K74913225-001-14-0 | HCRKCZRJWPKOAR-JTQLQIEISA-N | brinzolamide | 68844 | CA5A | trt | nan | CCN[C@H]1CN(CCCOC)S(=O)(=O)c2sc(cc12)S(N)(=O)=O |
| 6 | BRD-K94342292-001-01-3 | MDKAFDIKYQMOMF-UHFFFAOYSA-N | NS-11021 | 24825677 | KCNMA1 | trt | nan | FC(F)(F)c1cc(NC(=S)Nc2ccc(Br)cc2-c2nnn[nH]2)cc(c1)C(F)(F)F |
| 7 | BRD-K19975102-001-01-2 | YYDUWLSETXNJJT-MTJSOVHGSA-N | GNF-5837 | 59397065 | NTRK1 | trt | nan | Cc1ccc(NC(=O)Nc2cc(ccc2F)C(F)(F)F)cc1Nc1ccc2c(NC(=O)\C2=C/c2ccc[nH]2)c1 |
| 8 | BRD-K25244359-066-02-6 | WPEWQEMJFLWMLV-UHFFFAOYSA-N | apatinib | 11315474 | CSK | trt | nan | O=C(Nc1ccc(cc1)C1(CCCC1)C#N)c1cccnc1NCc1ccncc1 |
| 9 | BRD-K18131774-001-18-4 | OUZWUKMCLIBBOG-UHFFFAOYSA-N | ethoxzolamide | 3295 | CA5A | trt | nan | CCOc1ccc2nc(sc2c1)S(N)(=O)=O |
I recommend using the load_data/final parquet dataframe to get a list of all the images downloaded on disk.
Then, you can use the metadata/complete_metadata.csv file to get the metadata for each image and merge it with the load_data/final dataframe, on the source, plate and well columns.
A line of the load_data/final dataframe is equivalent to a "site" of a well, there are around 6 sites per well.
And a site is constituted of 5 images, one for each channel.
A line therefore corresponds to 5 images, one for each channel, in the columns FilePath_Orig{DNA,AGP,ER,Mito,RNA} and the columns Metadata_{Source,Batch,Plate,Well,Site} contain the metadata for the site.
The load_data/load_data_with_samples can be used if you want to look at the images that were not included in the local dataset.
The jobs folder is totally useless after the dataset has been downloaded.
Clone the code from GitHub:
git clone https://github.com/gwatkinson/jump_download.git
cd jump_downloadUse Poetry to install the Python dependencies (via pip). This command creates an environment in a default location (in ~/.cache/pypoetry/virtualenvs/). You can create and activate an environment, poetry will then install the dependencies in that environment:
poetry install --without dev # Install the dependencies
POETRY_ENV=$(poetry env info --path) # Get the path of the environment
source "$POETRY_ENV/bin/activate" # Activate the environmentThis project uses hydra to manage the configuration files. The configuration files are located in the conf folder.
There are 4 groups of configuration files:
- output_dirs: The output directories for the metadata, load data, jobs and images.
- filters: The filters to apply to the images, in order to reduce the number of images to download at source, plate and well level.
- processing: The processing to apply to the images during the download. This points to the main download class (that can be changed to modify the processing).
- run: The parameters related to the download script, such as the number of workers, whether to force the download, etc...
You can create new config files in each group, either by creating a new one from scratch or by overriding some options from the default config files.
By default, the file used to define the default groups is the conf/config.yaml file.
defaults:
- output_dirs: output_dirs_default # Default output_dirs group (defined in conf/output_dirs/output_dirs_default.yaml)
- filters: filters_default # Default filters group (defined in conf/filters/filters_default.yaml)
- processing: processing_default # Default processing group (defined in conf/processing/processing_default.yaml)
- run: run_default # Default run group (defined in conf/run/run_default.yaml)To change the default, you can either change the conf/config.yaml file so that it points to newly defined groups, change the default config of each group directly, or create a new root config and pass it to the commands with the -cn (or --config-name) argument.
See the local config for an example of a secondary config.
The download_example_script.sh script is an example of the commands needed to download the data. You probably should not use it as is, but rather copy the commands you need from it or create your own script.
# This script runs the commands in sequence to download the metadata and data files
CONF_NAME=$1
JOB_PATH=$2
echo "Downloading metadata and data files using the configuration file: $CONF_NAME"
echo "Creating Poetry environment"
poetry install --without dev # Install the dependencies
POETRY_ENV=$(poetry env info --path) # Get the path of the environment
source "$POETRY_ENV/bin/activate" # Activate the environment
echo "Downloading metadata files"
download_metadata -cn $CONF_NAME
echo "Downloading load data files"
download_load_data_files -cn $CONF_NAME
echo "Filtering and sampling images to download"
create_job_split -cn $CONF_NAME
echo "Downloading images using job in $JOB_PATH"
download_images_from_job -cn $CONF_NAME run.job_path=$JOB_PATH
# Or use the sub file
# condor_submit ./jump_download/condor/submit.sub
First, the metadata for the JUMP cpg0016 dataset can be found on github.com/jump-cellpainting/datasets. The script download_metadata (defined with poetry) gets the metadata and does some light processing (merge, ...):
download_metadata # -h to see helpThe main parameters that can be changed in the output_dirs group are:
metadata_dir
metadata_download_scriptThen, we need the load data files from the S3 bucket (see here to explore the bucket).
Those file contains the urls to download the images on S3 and are used to make the link with the metadata (merging by source, batch, plate and well).
This downloads all the load data files (around 216MB compressed but 11GB to download) and does some processing on it (merge the metadata, etc...)
resulting in a directory containing parquet files called /projects/cpjump1/jump/load_data/load_data_with_metadata by default:
download_load_data_filesThe main parameters that can be changed are:
load_data_dir
metadata_dir # Relies on the metadata downloaded in the previous stepThis is the next step, where we create the csv that are used to run the jobs on condor. This step also samples observations given rules set in the config.
Run the following command to create the job files:
create_job_splitThis script uses the load_data_with_metadata dataframe to create the job files.
It samples a number of images per well and applies source and plate level filters as well.
The output is a directory called /projects/cpjump1/jump/jobs by default.
It contains a ids folder with around 2100 parquet file that are all equivalent to one plate.
They have a similar format to the load_data_with_metadata dataframe but with only the selected observations, and with the additional columns:
output_dir: The directory where the images will be downloaded for that jobfilter: Whether the observation was filtered out or not (this should always beTrueas only the selected observations are kept)job_id: The id of the job (equivalent to the name of the csv file), the form is{source}__{batch}__{plate}
The function giving the output_dir and job_id are hard coded in the sample_load_data.py file. To modify the output paths you should change the functions create_output_dir_column and create_job_id_column.
The main parameters that can be changed are:
load_data_dir # Relies on the load data downloaded in the previous step
job_output_dir # The directory where the job files will be saved
source_to_disk # The mapping from source to disk (this is used to create the output_dir column)
plate_types_to_keep # The plate types to keep (this is used to filter images)
sources_to_exclude # The sources to exclude
# The number of observations to keep per well for each type of plate
{compound/orf/crispr/target}_number_of_{poscon/negcon/trt}_to_keep_per_wellThe resulting folder structure is:
* load_data/jobs/
* ids/
* {plate}__{batch}__{source}.parquet # The job files, one per plate, and the dropped images are not included
* submission.csv # The csv file that contain the path to all the job files that should not be droppedThe original load_data file is split into many subfiles in order to use them with Condor.
If you prefer downloading all the images at once, you can just use the load_data/jobs/ids parquet directory directly, which can loaded directly with pandas as a single dataframe.
Finally, the images can be downloaded using the following command:
download_images_from_csv run.job_path=<PATH><PATH> can be the path to a single parquet job file, or the directory containing all the job files (in which case all the job files will be downloaded).
The important parameters are:
download_class:
_target_: jump_download.images.final_image_class.Robust8BitCropPNGScenario # Points to the main download class
percentile: 1.0 # Arguments passed to the download class
min_resolution_x: 768
min_resolution_y: 768This script uses the Robust8BitCropPNGScenario.
It can be modified to use different parameters, or to use a different scenario.
The main class used to download and apply the processing to the images is the Robust8BitCropPNGScenario class.
It inherits from the BaseDownload class, which is a class that defines a framework to run a function on a list of jobs. The code is in the jump_download/base_class.py file.
The BaseDownload class is the abstract class that define a framework to download the data. The code is in the download/base_class.py file, and is used in the images and load_data_files modules.
flowchart TD
subgraph "BaseDownload Class"
direction LR
subgraph "download"
direction LR
a["get_job_list
#8594; list[dict]"]
c["post_process
list[dict] #8594; pd.DataFrame"]
subgraph b["download_objects_from_jobs"]
direction TB
ba["multiprocess_generator
list[dict] #8594; list[dict]"]
bb["execute_job
dict #8594; str"]
bb ~~~ ba
end
a --> b --> c
end
d("download_timed
@time_decorator(download)
")
end
The get_job_list, execute_job and post_process methods are abstract, and need to be implemented in the child class.
From there, the GetRawImages class is a child class of the BaseDownload class. It is used to download the raw images from the S3 bucket.
It has the following structure:
flowchart TD
subgraph "GetRawImages Class"
direction TB
subgraph init["#95;#95;init#95;#95;"]
direction TB
a(["global s3_client"])
b(["load_data_df"])
f(["max_workers"])
e(["force"])
c(["channels"])
d(["bucket_name"])
end
get_job_list["get_job_list
load_data_df#8594;list#91;dict#91;dict#93;#93;"]
execute_job["execute_job
dict#91;dict#93;#8594;str"]
init ~~~ get_job_list & execute_job
end
It implements the get_job_list and execute_job methods. The jobs are dictionaries with the following structure :
- source: Information on the image
- batch
- plate
- well
- the 5 channels as keys, the value being a sub dictionary :
- channel: the name of the channel (duplicate)
- s3_filename: the name of the file on the S3 bucket (Key for boto3)
- bucket_name: the name of the bucket (Bucket for boto3)
- buffer: the BytesIO object empty
The execute_job method uses the download_fileobj from boto3 to add the bytes object to the job dictionaries.
Finally, the Robust8BitCropPNG class is a child class of the GetRawImages class. It is used to download the raw images from the S3 bucket, and then process them to create the 8-bit PNG images.
flowchart TD
subgraph "Robust8BitCropPNGScenario Class"
direction TB
subgraph init["#95;#95;init#95;#95;"]
direction TB
a{{"GetRawImages class with parameters"}}
b(["percentile"])
c([min_resolution_x])
d([min_resolution_y])
e([out_dir])
f([out_df_path])
g([output_format])
end
subgraph get_job_list["get_job_list"]
direction LR
aa{{"GetRawImages.get_job_list()"}}
ab["Add file output paths"]
ac["And check if exist"]
aa --> ab --> ac
end
subgraph execute_job["execute_job"]
direction LR
ba{{"GetRawImages.execute_job()"}}
subgraph bb["job_processing"]
direction TB
bba["Load image from BytesIO"]
bbb["Apply image_processing <br> Crop + Scale + 8-bit"]
bba --> bbb
end
bc["Save to out_dir <br> with output_format"]
ba --> bb --> bc
end
subgraph post_process["post_process"]
direction LR
ca["Check that all images were <br> downloaded successfully"]
cb["Keep output path <br>and source/plate/well/site"]
cc["Save dataframe to out_df_path"]
ca --> cb --> cc
end
init ~~~ get_job_list
get_job_list --> execute_job
execute_job --> post_process
end
You can create your own class by inheriting from the BaseDownload class, and implementing the get_job_list, execute_job and post_process methods. It requires the following arguments:
- load_data_df: The path to the load data dataframe (defined in the config files)
- out_dir: The path to the output directory (usually the folder where the images will be saved, defined in the config)
- out_df_path: The path to the resulting dataframe
- max_workers: The number of workers to use for the multiprocessing (passed to the BaseDownload class)
- force: Whether to force the download of the images, even if they already exist
If you create your own class, you will also probably need to modify the main function in the jump_download/images/final_image_class.py module.
If you recreate it, the required arguments listed above are not mandatory (do as you wish).
This is the last step of the pipeline. It uses the download_plate command to download the images from the S3 bucket.
It consists only of a single .sub file that tells how much ressources should be used, and how many jobs should be run in parallel on different nodes.
condor_submit jump_download/condor/submit.subThis requires some of the previous steps, namely:
download_metadata
download_load_data_files
create_job_splitThis step can be ignored if you are not using HTCondor.
However, if you use it, change the content of the submit.sub file to match your needs.
# 1 - Describes the working directory of the job.
InitialDir = <path/to/project>
request_cpus = 8
request_memory = 2GB
MY.MaxThreads = 64
accounting_group = LongJob.Weekly
MY.Limit = (10000 / $(MY.MaxThreads)) * $(request_cpus)
concurrency_limits_expr = StrCat(MY.Owner,"_limit:", $(MY.Limit))
# 2 - Describes the program and arguments to be instantiated.
executable = <path/to/project>/jump_download/condor/download_plate.sh
arguments = $(job_csv_file) $(request_cpus)
# 3 - Describes the output files of the job.
output = <path/to/log/folder>/$(Cluster)-$(Process).output
error = <path/to/log/folder>/$(Cluster)-$(Process).error
log = <path/to/log/folder>/$(Cluster)-$(Process).log
stream_output = True
stream_error = True
notify_user = <email>
notification = Complete
# - Insert this job into the queue!
queue job_csv_file from <path/to/load_data_dir>/jobs/submission.csv