Link to Paper: "Pruning by Explaining: A Novel Criterion for Deep Neural Network Pruning" Also see our code and experiments on large image databases, VGG networks and ResNets: https://github.com/seulkiyeom/LRP_Pruning
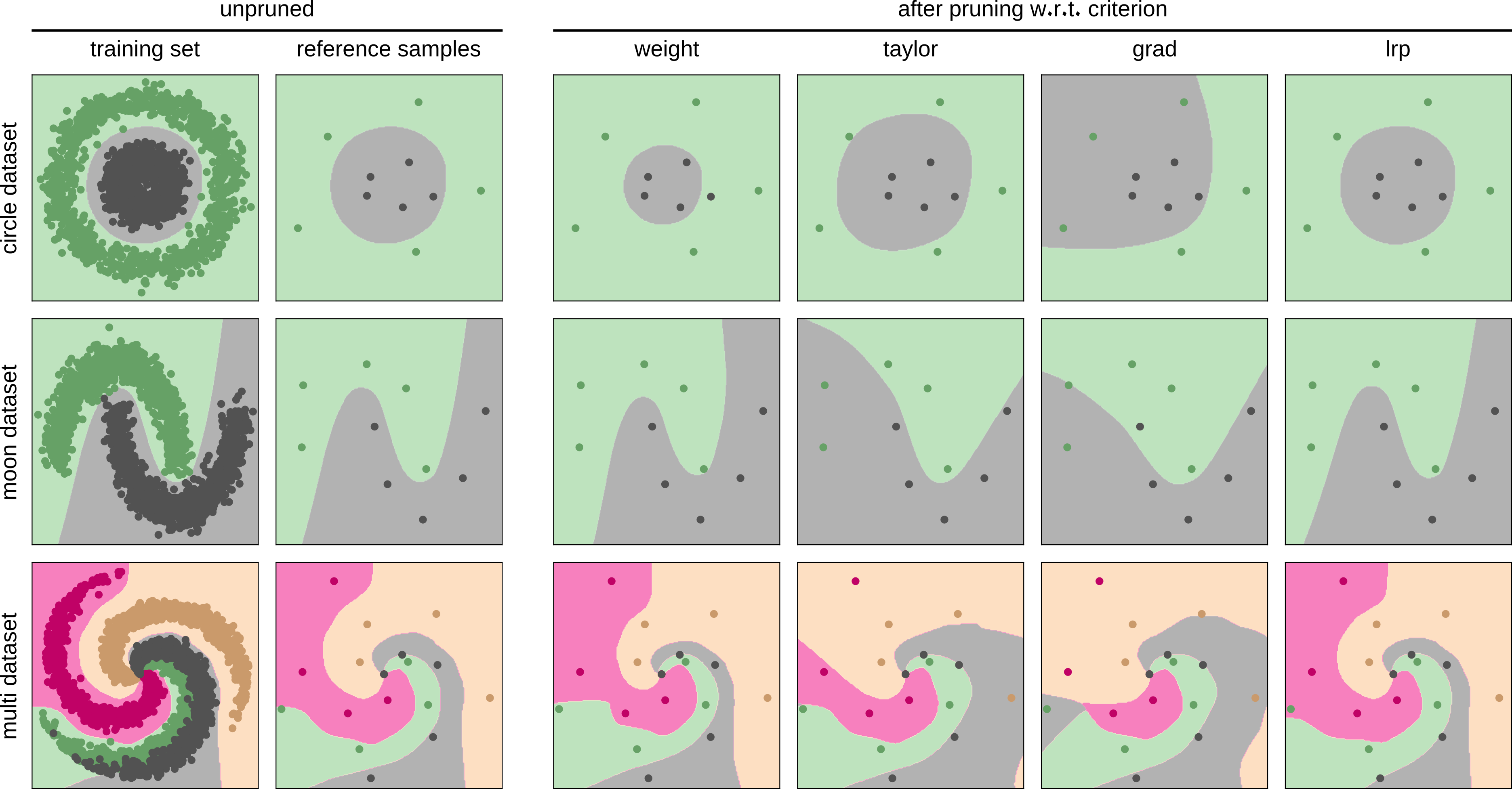 Qualitative comparison of the impact of the pruning criteria on the models’ decision function on three toy datasets.
1st column: scatter plot of the training data and decision boundary of the trained model,
2nd column: data samples randomly selected for computing the pruning criteria,
3rd to 6th columns: changed decision boundaries after the application of pruning w.r.t. different criteria.
Qualitative comparison of the impact of the pruning criteria on the models’ decision function on three toy datasets.
1st column: scatter plot of the training data and decision boundary of the trained model,
2nd column: data samples randomly selected for computing the pruning criteria,
3rd to 6th columns: changed decision boundaries after the application of pruning w.r.t. different criteria.
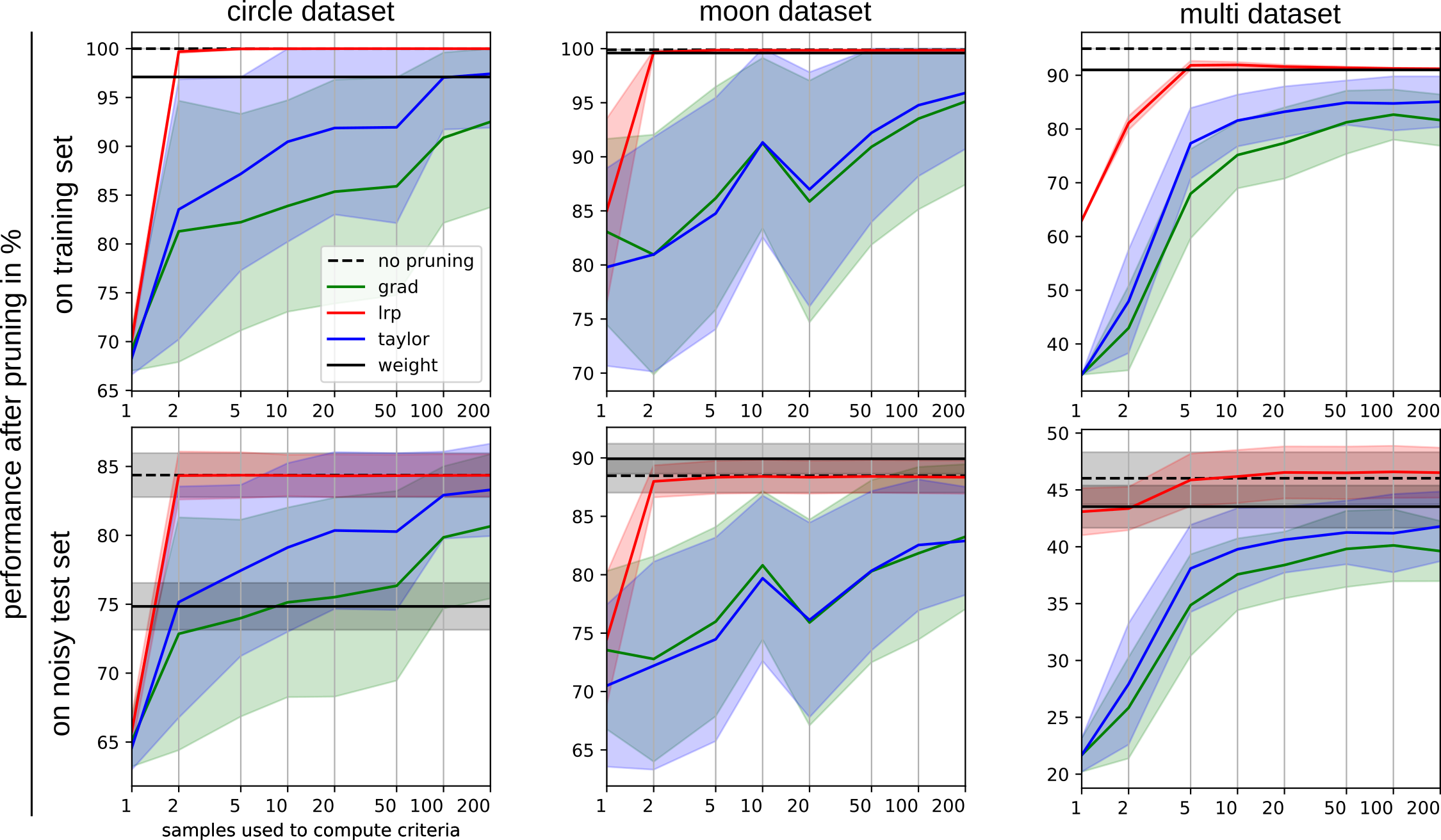 Pruning performance comparison of criteria depending on the number of reference samples per class used for criterion computation.
1st row: Model evaluation on the training data.
2nd row: Model evaluation on an unseen test dataset with added gaussian noise (
Pruning performance comparison of criteria depending on the number of reference samples per class used for criterion computation.
1st row: Model evaluation on the training data.
2nd row: Model evaluation on an unseen test dataset with added gaussian noise (), which have not been used for the computation of pruning criteria.
Columns: Results over different datasets.
Solid lines show the average post-pruning performance of the models pruned w.r.t. to the evaluated criteria
"weight" (black), "taylor" (blue), "grad(ient)" (green) and "lrp" (red) over 50 repetitions of the experiment.
The dashed black line indicates the model's evaluation performance without pruning.
Shaded areas around the lines show the standard deviation over the repetition of experiments.
If you make use of, or are inspired by our work and code, please cite our paper
@article{yeom2021pruning,
title={Pruning by explaining: A novel criterion for deep neural network pruning},
author={Yeom, Seul-Ki and
Seegerer, Philipp and
Lapuschkin, Sebastian and
Binder, Alexander and
Wiedemann, Simon and
M{\"u}ller, Klaus-Robert and
Samek, Wojciech},
journal={Pattern Recognition},
pages={107899},
year={2021},
publisher={Elsevier}
}
First, familiarize yourself with the command line options.
$ python main.py --help
usage: main.py [-h] [--dataset DATASET] [--criterion CRITERION]
[--numsamples NUMSAMPLES] [--seed SEED]
[--rendermode RENDERMODE] [--colormap COLORMAP]
[--logfile LOGFILE] [--generate] [--analyze] [--ranklog]
[--noisytest NOISYTEST]
Neural Network Pruning Toy experiment
optional arguments:
-h, --help show this help message and exit
--dataset DATASET, -d DATASET
The toy dataset to use. Choices: moon, circle, mult
--criterion CRITERION, -c CRITERION
The criterion to use for pruning. Choices: lrp,
taylor, grad, weight
--numsamples NUMSAMPLES, -n NUMSAMPLES
Number of training samples to use for computing the
pruning criterion.
--seed SEED, -s SEED Random seed used for (random) sample selection for
pruning criterion computation/testing.
--rendermode RENDERMODE, -r RENDERMODE
Is result visualization desired? Choices: none, svg,
show
--colormap COLORMAP, -cm COLORMAP
The colormap to use for rendering the output figures.
Must be a valid choice from matplotlib.cm .
--logfile LOGFILE, -l LOGFILE
Output log file location. Results will be appended.
File location (folder) must exist!
--generate, -g Calls a function to generate a bunch of parameterized
function calls and prepares output locations for the
scripts. Recommendation: First call this tool with
"-g", then execute the generated scripts. If
--generate is passed, the script will only generate
the scripts and then terminate, disregarding all other
settings.
--analyze, -a Calls a function to analyze the previously generated
log file. If --analyze is passed (but not --generate)
the script will analyze the log specified via
--logfile and draw some figures or write some tables.
--ranklog, -rl Triggers a generation of scripts (when using -g), and
an evaluation output and analysis (when using -a) for
neuron rank corellations and and neuron set
intersection.
--noisytest NOISYTEST, -nt NOISYTEST
The -nt parameter specifies the intensity of some
EXTRA gaussian noise added to the dataset. That is,
given the parameter >0, a secondary larger test set
will be generated just for the purpose of testing the
model (not for pruning).
Now, in order to (re)create our results, you can either (re)generate the our toy experiment scripts using the --generate (-g) switch.
This will then generate shell scripts to replicate the full pruning experiment. You will find these scripts are pre-generated in scripts.
Optionally consider adding the --ranklog (-rl) switch to generate shell scripts to only analyze the neuron selection for prunining per criterion (without performing the full prunig procedure, but spamming the log with megabytes worth of pruning order information instead), reference sample size and random seed for each toy dataset. The pre-generated scripts for the latter scenario can be found in scripts-rankanalysis.
$ python main.py -g [-rl]
Thereafter, you can execute the experiments by running the generated shell scripts, e.g.
for s in $script_dir/*.sh ;
do
bash $s & # consider removing the "&" if your machine is lacking the RAM and CPU cores for parallel execution
done
Outputs will be written into a plain text log file specified via the --logfile (-l) during script generation
pre-computed result logs containing the results from our paper can be found.
Logs pre-computed for full pruning runs over 50 random seeds, as well as neuron rank analysis logs, can be found in ./output/log.txt and output-rankanalysis./log.txt respectively.
A shell script as well as a resulting log file and csv figure files for generating the qualitative toy figure toy-figure-mk2-test.png above can be found in folder output-toy-figure-mk2.
The log files for comparing model performances (resulting i.e. in the pruning performance figure output/combined-processed.png above) can be analyzed by adding the switch --analyze (-a), which overrides any other instructions (except for -g, which overrides the behavior of -a). An optional addition of the --rl switch directs the analysis towards neuron selection during pruning, instead of performance a comparison. An analysis can be instigated e.g. via specification of the log file containing the various statistics
$ python -a -rl -l output-rankanalysis/log.txt
Corresponding analysis outputs (i.e. large amounts of tables in plain text files when using the -rl switch, or line plots in svg file format when analyzing pre and post-pruning model performance) will be created next to the log file selected for analysis, and can already be found precomputedly in the output* folders of this repository.