EEG preprocessing for DEAP dataset
This repo is based on DEAP, which is among the most-cited datasets for EEG-based emotion recognition. You can find scripts to a) download DEAP, b) preprocess its EEG signals and c) perform feature extraction (Power Spectral Density, Differential Entropy, Power Spectral Density asymmetry, Differential Entropy asymmetry).
Note that, to download DEAP dataset using this code, you need to create a JSON file containing your credentials, using the create_json.py file.
After downloading the dataset in BDF file format, you can proceed to preprocessing. The pipeline for EEG preprocessing is based on the well-known steps of Steve Luck. More specifically, the preprocessing script performs the following steps:
- Load .bdf file for each subject
- Get channel names (1-32 are EEG, 33-40 are EMG/EOG, the last one [48/49] is Stimuli status)
- Drop non-EEG channels (keep stimulus/status channel)
- Set montage of electrode locations to Biosemi32
- Get sampling frequency
- Apply filters on data: notch filter @ 50Hz, bandpass filter @ 4-45Hz
- Re-reference EEG channels to the common average reference
- Get events from stimulus/status channel
- Keep only trial-start events (event ID == 4 is the stimulus onset, i.e. the beginning of each trial)
- Epoch data, using the trial-start events, and setting tmin=-5.0 and tmax=+60.0
- Define an ICA transformation, using FastICA and 32 channels
- Fit the ICA to the epoched data 12a: Plot ICA sources (optional) 12b: Save ICA sources as figures (optional)
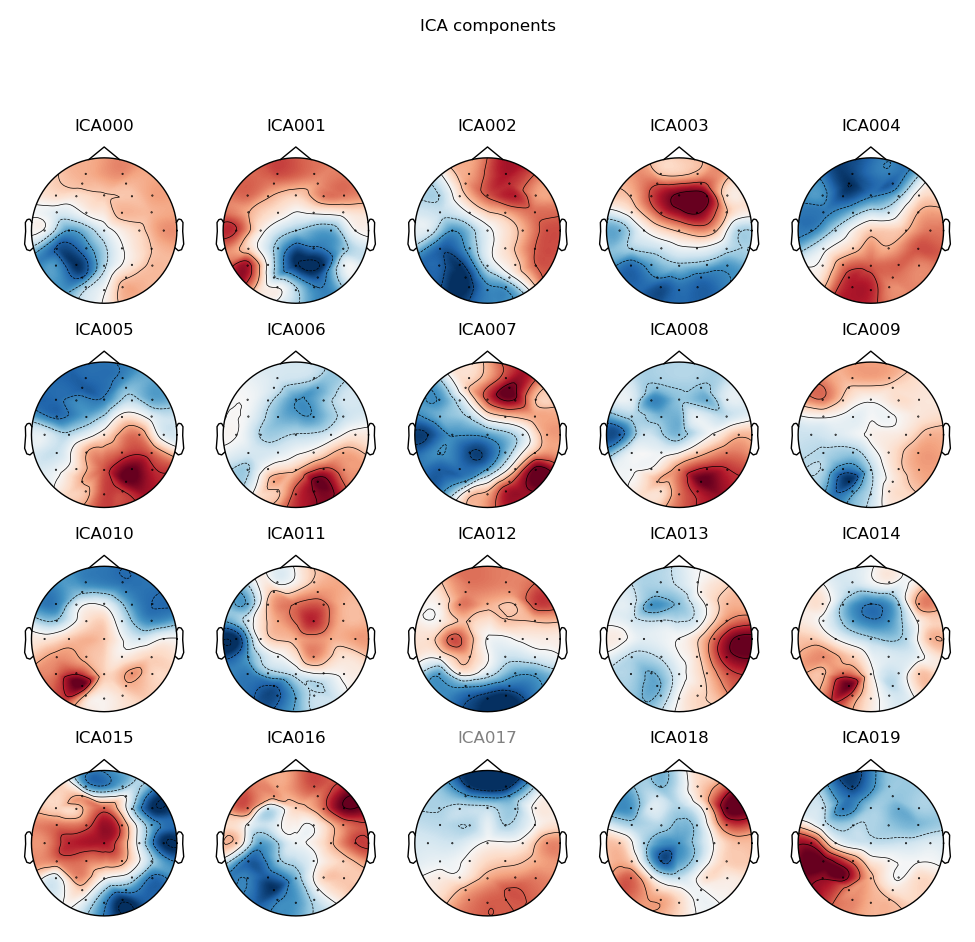
- Plot ICA components and manually reject eye-movement related components 13a: Save ICA components as figures
- Apply the fitted ICA to the epoched data, to obtain the cleaned epoched data
- Plot the PSD of the clean epoched data
- Downsample the frequency of the cleaned epoched data, from 512Hz to 128Hz
- Get the EEG time-series of the downsampled cleaned epoched data
- Re-order the EEG channels, to follow the Geneva order
- Re-order the trials, to follow the experiment_ID order (same video stimulus order)
You can see an example of a subject's ICA components here:
You can see an example of a subject's PSD plot here: