ProteinLearn makes complex protein data understandable for diverse user groups, including students, drug designers, and researchers.
-
AlphaFold, ESM, and other protein LLMs have made significant progress in predicting 3D structures given amino acid sequences. However, they are unable to provide human-understandable language to further explain the functionality of the protein.
-
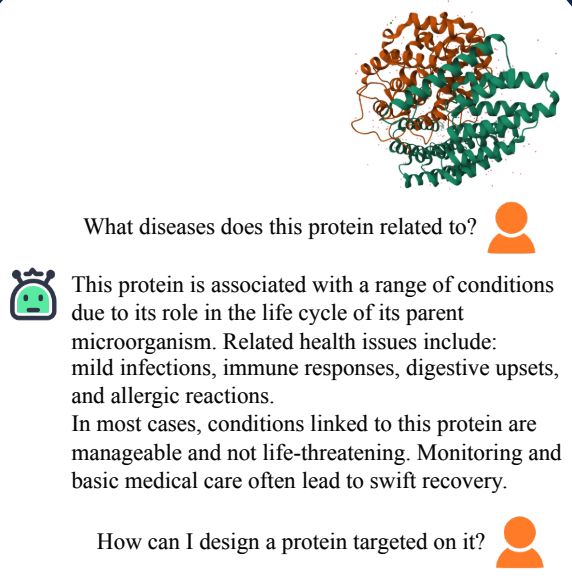
ProteinChat has been proposed to help us understand protein structures through natural language. However, it still suffers from hallucination when encountering proteins not included in the training dataset.
-
This problem can be circumvented by introducing hallucination-free LLMs from the
Hallucinations Leaderboardor by using a RAG system.
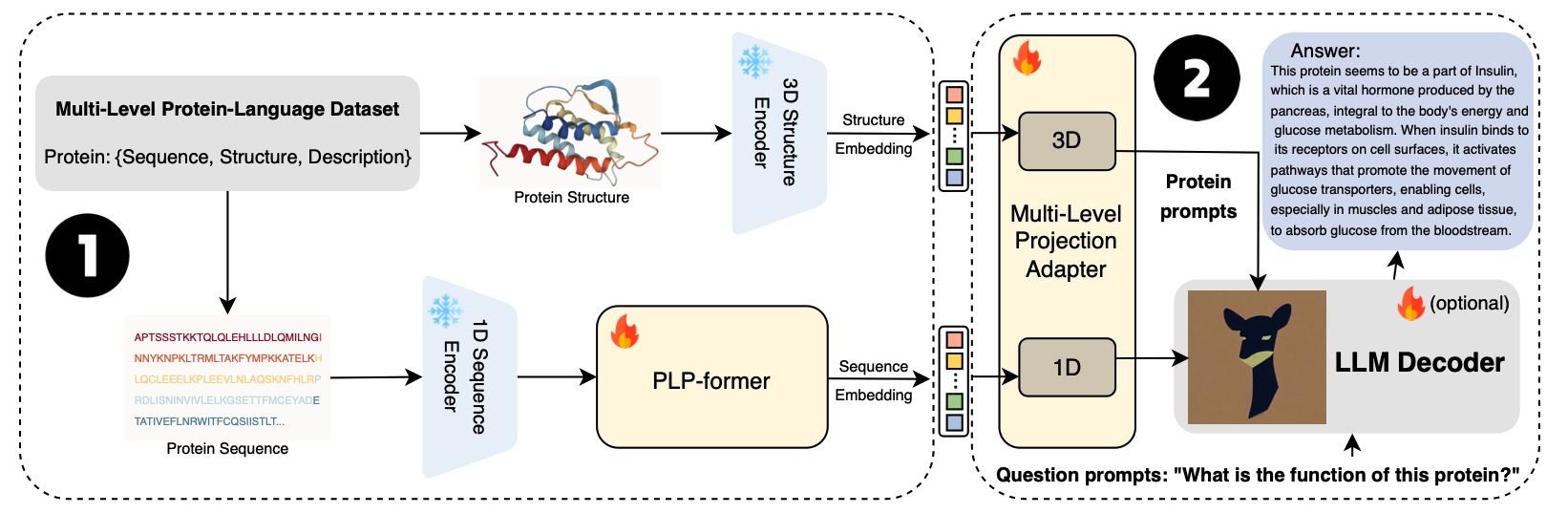
- Amino acid squences together with the proteins' 3D structure are encoded together to provide comprehensive information.
- The adapter is trained to form soft prompt as input for the downstream LLMs to generate explanation
- (0) Project discovery
- (1) Explore protein data sources for evaluation and RAG
- (2-4) Reseach and design ProteinLearn pipeline
- (5-6) Experiment LLMs on
Hallucinations Leaderboard - (6-7) Experiment RAG with external protein data sources
- (8) Build user interface and 3D protein visualization
- (9) Ensure CI/CD is robust and dockerizes
- (10) Documentation and Clean up