Wrapping some of AmberTools's programs for Jupyter notebook. For now, this is only for fun and you should not rely on this unstable API. Hopefully it's still useful.
Install
git clone https://github.com/hainm/jamber
cd jamber
python setup.py install
# If using ambertools
# amber.python setup.py installRequire
- AmberTools >= 16
- nglview (optional): For visualization in Jupyter notebook
Example
-
leap
from jamber import leap
command = """
source leaprc.protein.ff14SB
seq = sequence {ALA ALA ALA}
saveamberparm seq seq.prmtop seq.rst7
"""
leap.run(command)- builder
from jamber.builder import build_bdna
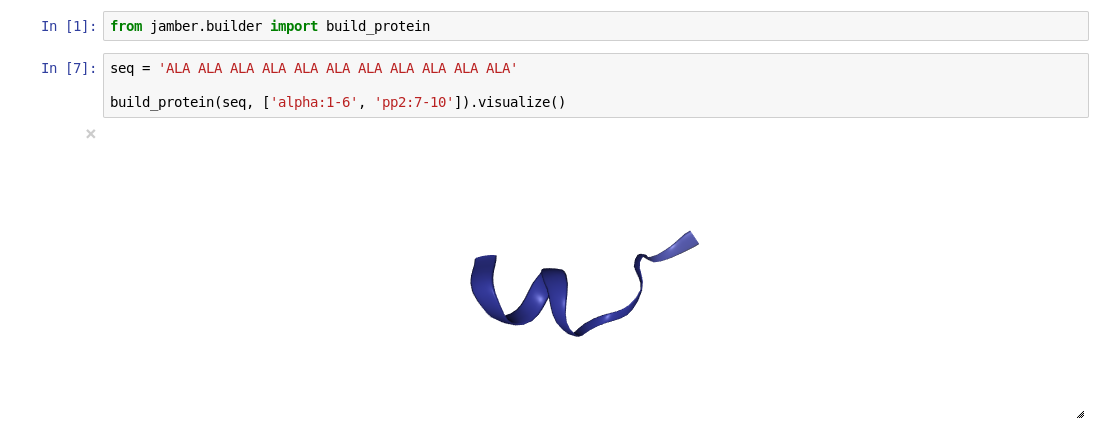
build_bdna(seq='AAAAAA')- build protein with given secondary structure