have a look at DepMap
What you need to process the Quarterly DepMap-Omics releases from Terra.
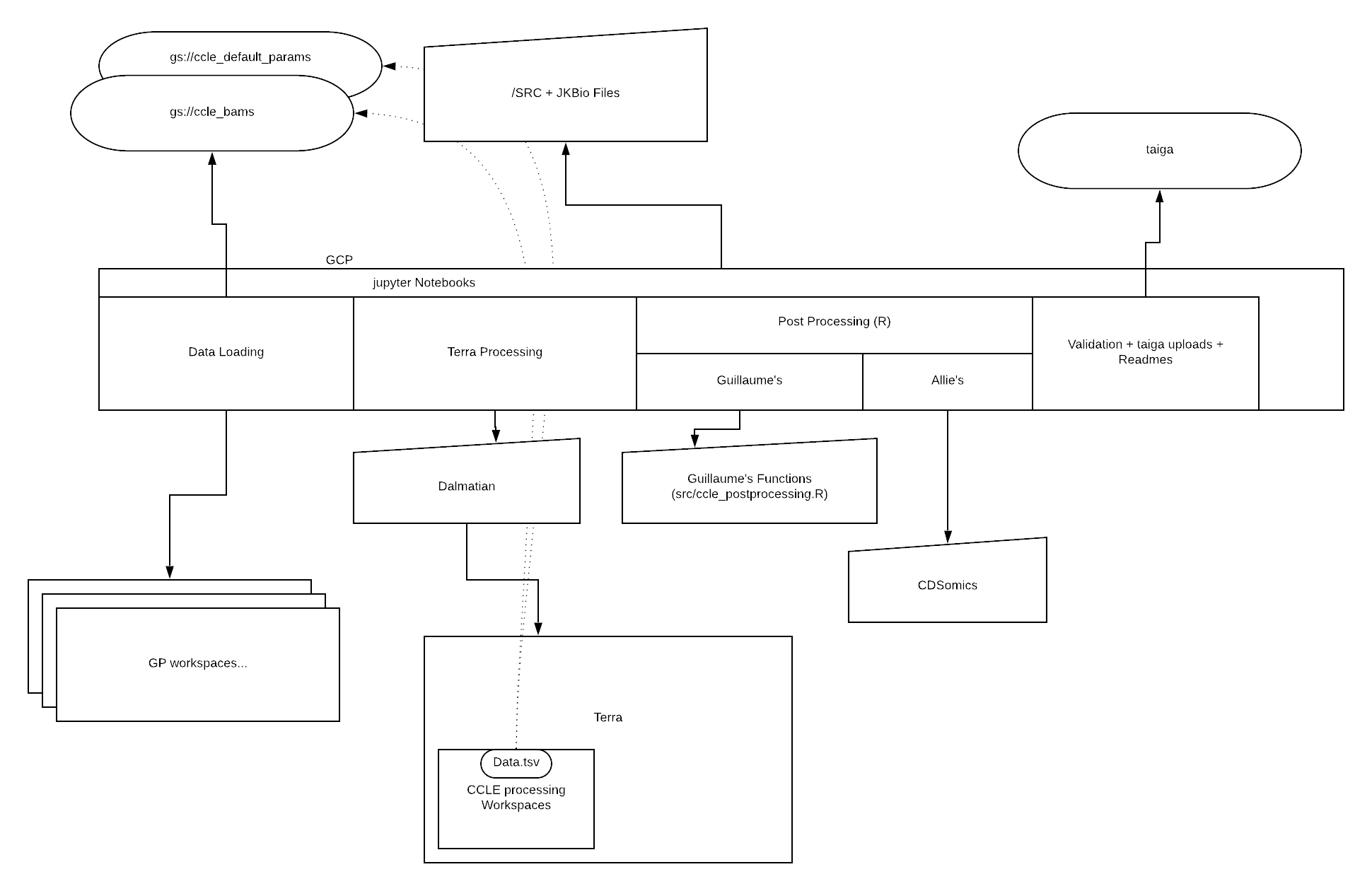
Here is a presentation of the pipeline.
If you are not familiar with these notions, we will first recommend you get more knowledge into each:
git clone http://github.com/BroadInstitute/depmap_omics.git && cd depmap_omics
pip install -e .
Some important data and code from the genepy Library.
Use the instructions in the genepy page to install the package.
- You will need to install jupyter notetbooks and google cloud sdk
- install Google Cloud SDK.
- authenticate my SDK account by running
gcloud auth application-default loginin the terminal.
-
and GSVA for ssGSEA in R
RrunR -e 'if(!requireNamespace("BiocManager", quietly = TRUE)){install.packages("BiocManager")};BiocManager::install(c("GSEABase", "erccdashboard", "GSVA", "DESeq2"));' -
For Python use the requirements.txt file
pip install -r requirements.txt
To learn about the tools we use in the pipeline, see here for a detailed walkthrough
- You will need to request access to the following terra workspaces:
The current owners of these workspaces should give you access.
- For the mutation pipeline you will also need to request dbGaP access (required for TCGA workflows). See CCLE/new hiree section on Asana for details.
- Acquire access to required billing projects (e.g. broad-firecloud-ccle). See CCLE/new hiree section on Asana for details.
- Get access to the following Terra groups:
- depmap_ccle_data
- depmap-pipelines
- ccle-pipeline
- If you need to get access to the data delivered by GP, use the following links:
- Request access to the data bucket
gs://cclebams/ - You will need also access to the billing project
broad-firecloud-ccle
- In order to access and upload data, you will need to login to taiga with your broad google account and set up your token.
- In order to run the imports gsheets, you need to make sure your broad google account has access to the cell line info sheets. In addition, you will need to obtain the following google API credential files:
- Go to console, acquire access if you don't have it already. Under credentials -> create credentials -> OAuth Client ID -> application type = Desktop app -> create. Download
client_secreat.jsonand save as~/.client_secret.json(Refer to quickstart here). - After calling
gsheets.from_filesfor the first time using~/.client_secret.json, log in via google following the prompt in browser,storage.jsonwill be created automatically. Save it as~/.storage.json. - Follow instruction here, create service account if needed, and save key file as
../.credentials.json.
- Go to console, acquire access if you don't have it already. Under credentials -> create credentials -> OAuth Client ID -> application type = Desktop app -> create. Download
*Remember to share relevant gsheets with the service account (client_email in ../.credentials.json).
We are instantiating all the parameters needed for this pipeline to run
We are looking for new samples in a range of workspaces.
They are quite messy and might contain duplicates and/or broken file paths...
- We are thus looking at the bam files one by one and comparing them with bams we have onboarded.
- We remove broken files, duplicates and add new version of a cell line's bam if we find some.
This involves comparing to the list in the Google sheet "Lines to Release"
As the list cannot be parsed, we are not comparing it for now
To learn about the tools we use in the pipeline, see here for a detailed walkthrough
- You first need a Terra account with correct billing setup. See here for a tutorial on getting started.
- If you haven't already, create a workspace under the billing project of your choice. If you need to process both RNA and WGS data, we recommend creating one workspace for each.
- Import the WDL scripts by following the links to dockstore and clicking on launch with terra (note: you'll need both pipeline and aggregate for each data type):
- DepMap's workspace configurations are saved after each data release under
data/. We recommend using configurations from the latest quarter. For example, if the latest release is21Q4, you should be able to find the configurations indata/21Q4/RNAconfig/all_configs.jsonanddata/21Q4/RNAconfig/all_configs.jsonfor RNA and WGS, respectively. - Set up the right inputs and outpus for your workflows according to
inputs_[WORKFLOW_NAME].jsonandoutputs_[WORKFLOW_NAME].jsonfiles, which are under the same directory asall_configs.json. - Load your samples so that their bam and bam index google storage filepath get listed in the right data column to WGS_pipeline and RNA pipeline.
- Create a sample set with the set of samples you want to analyse. Make sure the name of this sample set on terra is the same as
SAMPLESETNAMEinconfig_prod.py.
Once this is done, you can launch your jupyter notebook server and run the *_CCLE jupyter notebooks corresponding to our RNA pipeline and WGS pipeline (older versions for WES (CN and mutations are available in a previous commit labelled 20Q4)).
Remark:
- you will need to use the
postProcesssing()functions for post processing instead of the CCLE ones in theccle.pymodule. - you will need to change some of the variables in the
config_prod.py. - you won't be able to run the function conditional on the CCLE boolean. You can however reimplement them to create your own pipeline.
The notebook architectures are as follows:
- Uploading and preprocessing
- Running the Terra Pipelines
- Downloading and postprocessing the samples
To run the CCLE pipeline we follow the installation process above and then boot up a GCP instance to run the notebooks from it.
You can find more documentation about the range of workspaces that have been created: here
We are using a set of key tools to process the sequencing output:
- star:
- rsem:
- star fusion:
- mutect:
- mutect2:
- gatk cnv:
- strelka:
The following flowchart provides another good overview of what the pipeline is doing.
What is explained below comes from the notebook's documentations and might be better understood by reading them directly on the notebooks
The first phase really is about getting samples generated at the broad and located into different places. Looking for duplicates and finding/adding the metadata we will need in order to have coherent and complete sample information. For external users, this is not something you would need to run. Please skip directly to part2.
Remarks:
- in the initialization step, external users might want to remove any import related to
taigaandgsheetto not cause any errors. - feel free to reuse
createDatasetWithNewCellLines,GetNewCellLinesFromWorkspacesor any other function for your own needs.
We are using Dalmatian to send requests to Terra, so before running this part, you need to make sure that your dalmatian WorkspaceManager object is initialized with the right workspace you created and that the functions take as input you workflow names. You also need to make sure that you created your sample set with all your samples and that you initialized the sampleset string in config_prod.py with its name.
You can then run the following two pipelines on your samples. The whole process may take 3-7 days.
We are running the following workflows in this order to generate copy number and mutation datasets:
WGS_pipeline imports and runs several sub-processes to generate copy number segments and MAF data.
WGS_aggregate aggregates CN segments and MAFs into their respective files.
The outputs to be downloaded will be saved under the sample set that you ran. The outputs we use for the release are:
-
combined_seg_file
-
filtered_CGA_MAF_aggregated
-
unfiltered_CGA_MAF_aggregated
-
Note that additional files are used for QC
We are generating both expression and fusion datasets with RNAseq data. Specifically, we use the GTEx pipeline to generate the expression dataset, and STAR-Fusion to generate gene fusion calls. This task also contains a flag that lets you specify if you want to delete the intermediates (fastqs) that can be large and might cost a lot to store. Run the following tasks on all samples that you need, in this order:
RNA_pipeline imports and runs several sub-processes to generate RNA expression and fusion data matrices.
RNA_aggregate aggregates expression and fusion data files into their respective aggregated file.
The outputs to be downloaded will be saved under the sample set that you ran. The outputs we use for the release are:
- rsem_genes_expected_count
- rsem_genes_tpm
- rsem_transcripts_tpm
- fusions_star
Finally, we save the workflow configurations used in the pipeline runs
Remarks:
- for the copy number pipeline we have parametrized both an XX version and an XY version, we recommend using the XY version as it covers the entire genome
- for the mutation pipeline we are working on Tumor-Normal pairs which explain some of the back and forth between the two workspace data table. (workflows works as well with or without matched normals.)
- for the expression pipeline, we have an additional set of workflows to call mutations from RNAseq, this might not be relevant to your need.
This step will do a set of tasks:
- clean the workspaces by deleting large useless files, including unmapped bams.
- retrieve from the workspace interesting QC results.
- copy realigned bam files to our own data storage bucket.
- download the results.
- remove all duplicate samples from our downloaded file (keeping only the latest version of each sample).
You would only be interested here at minima in the result downloading
...and postprocessing tasks. The main postprocessing steps for each pipeline are as followed:
copynumbers.py contains the main postprocessing function (postProcess() and their wrappers for internal use) responsible for postprocessing segments and creating gene-level CN files.
mutations.py contains postProcess() (and its wrappers for internal use, including one for filtered and one for unfiltered mutation data), which is responsible for postprocessing aggregated MAF files and generating various types of mutation datasets.
expressions.py contains the main postprocessing function (postProcess() and their wrappers for internal use) responsible for postprocessing aggregated expression data from RSEM, which removes duplicates, renames genes, filters and log transforms entries, and generates protein-level expression data files.
Functions responsible for postprocessing aggregated fusion data can be found in fusions.py. We want to apply filters to the fusion table to reduce the number of artifacts in the dataset. Specifically, we filter the following:
- Remove fusions involving mitochondrial chromosomes, or HLA genes, or immunoglobulin genes
- Remove red herring fusions (from STAR-Fusion annotations column)
- Remove recurrent in CCLE (>= 25 samples)
- Remove fusion with (SpliceType=" INCL_NON_REF_SPLICE" and LargeAnchorSupport="No" and FFPM < 0.1)
- Remove fusions with FFPM < 0.05 (STAR-Fusion suggests using 0.1, but looking at the translocation data, this looks like it might be too aggressive)
Remarks:
- in the RNAseq pipeline we have an additional sub-pipeline at the end of the notebook to process the fusion calls from starFusion
- to get the exact same results as in CCLE, be sure to run
genecn = genecn.apply(lambda x: np.log2(1+x))to the genecn dataframe in the CNV pipeline (present at the end of the validation steps). - we do not yet have integrated our germline calling in the mutation pipeline but you can still find the haplotypeCaller|DeepVariant workflows and their parameters
We then perform the following QC tasks for each dataset. These tasks should not be very interesting for external user as they revolve around manual checks of the data, comparison to previous releases, etc.
Once the CN files are saved, we load them back in python and do some validations, in brief:
- mean, max, var...
- to previous version: same mean, max, var...
- checkAmountOfSegments: flag any samples with a very high number of segments
- checkGeneChangeAccrossAll: flag any genes which stay at a similar value across all samples
Compare to previous release (broad only)
We compare the results to the previous releases MAF. Namely:
- Count the total number of mutations per cell line, split by type (SNP, INS, DEL)
- Count the total number of mutations observed by position (group by chromosome, start position, end position and count the number of mutations)
- Look at specific differences between the two MAFs (join on DepMap_ID, Chromosome, Start position, End position, Variant_Type). This is done for WES only
Overall the filters applied after the CGA pipeline are the following:
We remove everything that:
- has AF<.1
- OR coverage <4
- OR alt cov=1
- OR is not in coding regions
- OR is in Exac with a frequency of >0.005%
- except if it is either
- in TCGA > 3 times
- OR in Cosmic > 10 times
- AND in a set of known cancer regions.
- except if it is either
- OR exist in >5% of the CCLE samples
- except if they are in TCGA >5 times
Once the expression files are saved, we do the following validations:
- mean, max, var...
- to previous version: same mean, max, var...
- we QC on the amount of genes with 0 counts for each samples
After QC, we are also preparing the data to be released to different groups, removing the samples per access category: Blacklist|Internal|DepMapConsortium|Public.
We are then uploading the data to a server called taiga where it will be used in the depmap portal
For our 2 computation pipelines for depmap omics:
- Expression (RNA)
- Copy number and Mutations (WGS)
Each:
- can be run in a jupyter notebook file,
- gets data from Terra workspace's gcp buckets managed by Broad's Genomics Platform + DevOps,
- updates the sample TSVs on Terra with paths to the files,
- computes the results for each samples by running workflows,
- downloads the results and postprocesses them with additional local functions,
- performs QC and uploads them to taiga (internal only).
data/ Contains important information used for processing, including terra workspace configurations from past quarters
src Contains the location of function files
*_pipeline Contains some of the pipeline's workflows' wdl files and script files used by these workflows
ccle_tasks Contains a notebook for each of the different additional processing that the CCLE team has to perform as well as one-off tasks run by the omics team
legacy Contains the previous R markdown files that were used as templates for the previous pipeline's post-processing
readmes Contains some of the depmap readmes
temp Contains the temp file that can get removed after processing (should be empty)
documentation Contains some additional files for documenting the pipelines
for CN pons are made from a set of ~400 normals from the GTEx project as they were sequenced in the same fashion as CCLE samples with the same set of baits. you can see the ID of the samples in data/samples_for_pons.csv.
We have created pons for each bait set and using XY only.
We have used workflow from the pipeline:
gatk/CNV_Somatic_Panel_Workflow
The data we are presenting comes from different WES targets/baits/intervals.
We are currently using Illumina ICE intervals and Agilent intervals. you can find their related PON files and interval files as parameters in our workspace files in data/xQx.json
additional auxilliary data is used and listed in some of our workflow, like the CGA pipeline. You will be able to find them by looking at the wdl scripts of each pipelines and looking into the data/xQx.json` files for workspace data files.
@jkobject @gkugener @gmiller @5im1z @__BroadInsitute
For any questions/issues, send us an email at ccle-help@broadinstitute.org or write down an issue on the github repo