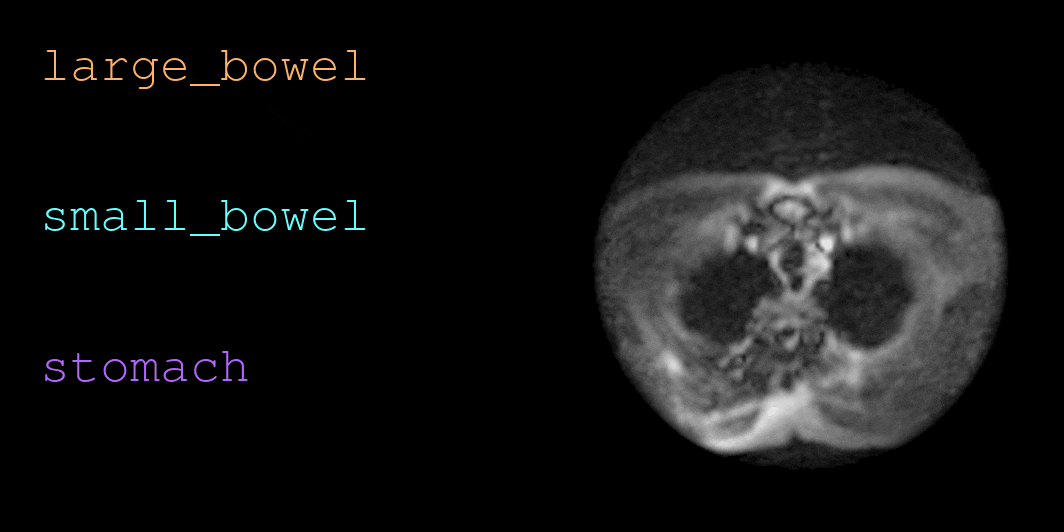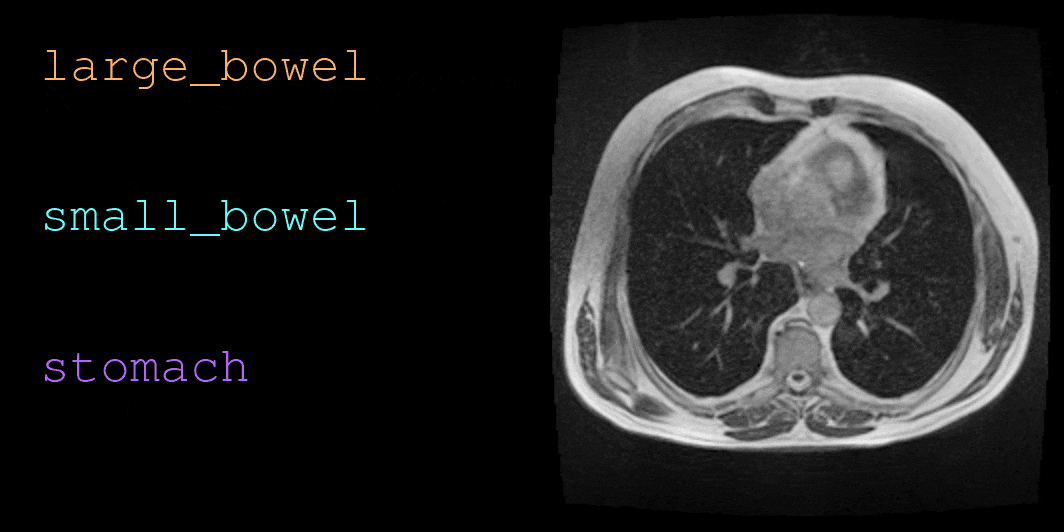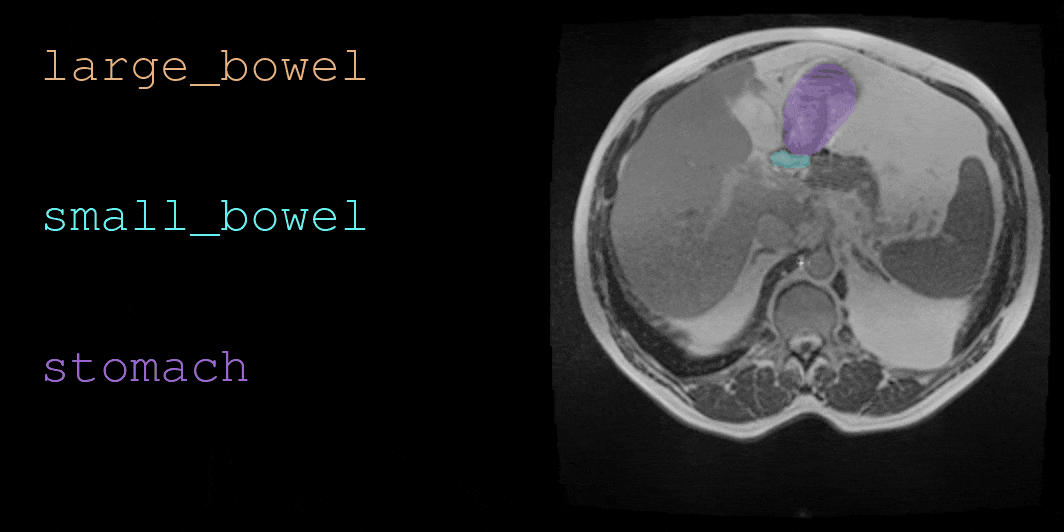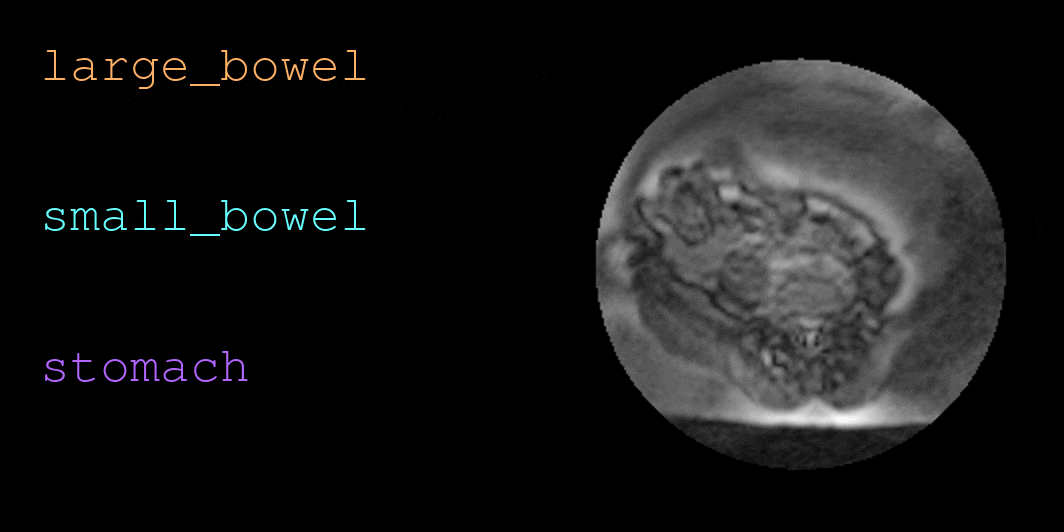This repository contains code for loading and visualizing MRI data and annotations used in UW-Madison GI Tract Image Segmentation challenge. See below for a sample visualization.
- 03/21/2023: Initial commit.
Follow INSTALL.md for installing necessary dependencies
UW-Madison GI Tract Image Segmentation Dataset
The MRI data and their annotations are hosted on Kaggle. A detailed description of the dataset and the downloading link can be found in this webpage.
Note: The training set can be directly downloaded, whereas the test set is only available when the code is submitted to Kaggle. For submission to Kaggle, please refer to instructions here.
Download and Unpack the Dataset
To use the code in this repository, please follow the steps to download and prepare the dataset.
- Download uw-madison-gi-tract-image-segmentation.zip from this webpage. A Kaggle account is needed.
- Unpack the zip file under ./data (or elsewhere and link to ./data).
- The folder structure should look like
This folder
│ README.md
│ ...
│
└───data/
│ └───train/
│ │ └───case2
│ │ └───case6
│ │ └───...
│ └───train.csv
│ ...
|
- To see how to use the code, run
python ./load_data.py --help- To load a single scan (e.g., case136_day25), run
python ./load_data.py ./data/train.csv ./data/train case136_day25- To visualize a single scan (e.g., case136_day25), run
python ./load_data.py ./data/train.csv ./data/train --vizAn animated GIF (e.g., case136_day25.gif) will be saved under ./outputs.
This repository also contains sample code for evaluating 3D Dice score and 3D Hausdorff distance. See common.py.
Yin Li (yin.li@wisc.edu)