BinCrisp
binCrisp is a simple script to detect CRISPR sequences from whole genome sequences using pilercr and produces visulisation matrix of CRISPR regions (in svg using svgwrite)
Dependencies include:
- Python 2.7+
- pilercr must be installed and on PATH: http://www.drive5.com/pilercr/
- svgwrite: https://pypi.python.org/pypi/svgwrite
Be sure these are installed and on your path. See USAGE for optional parameters. Easiest way to set up svgwrite is with pip:
pip install -r requirements.txt
binCrisp has one module (at the moment): findcr
findcr
findcr is the main module of the script. It requires a directory with all the genomes you wish to use and an file name to name all the output.
It combines all genomes sequences in input directory and it runs pilercr from the path. The results from pilercr are summarised and used to draw and presence absence matrix as svg using svgwrite:
python binCrisp.py findcr GenomeDir/ OutputFileName
The -d or --draw2 flag will space out CRISPR results into a grid to easily show presence/ absence:
python binCrisp.py findcr -d GenomeDir/ OutputFileName
binCrisp output will include;
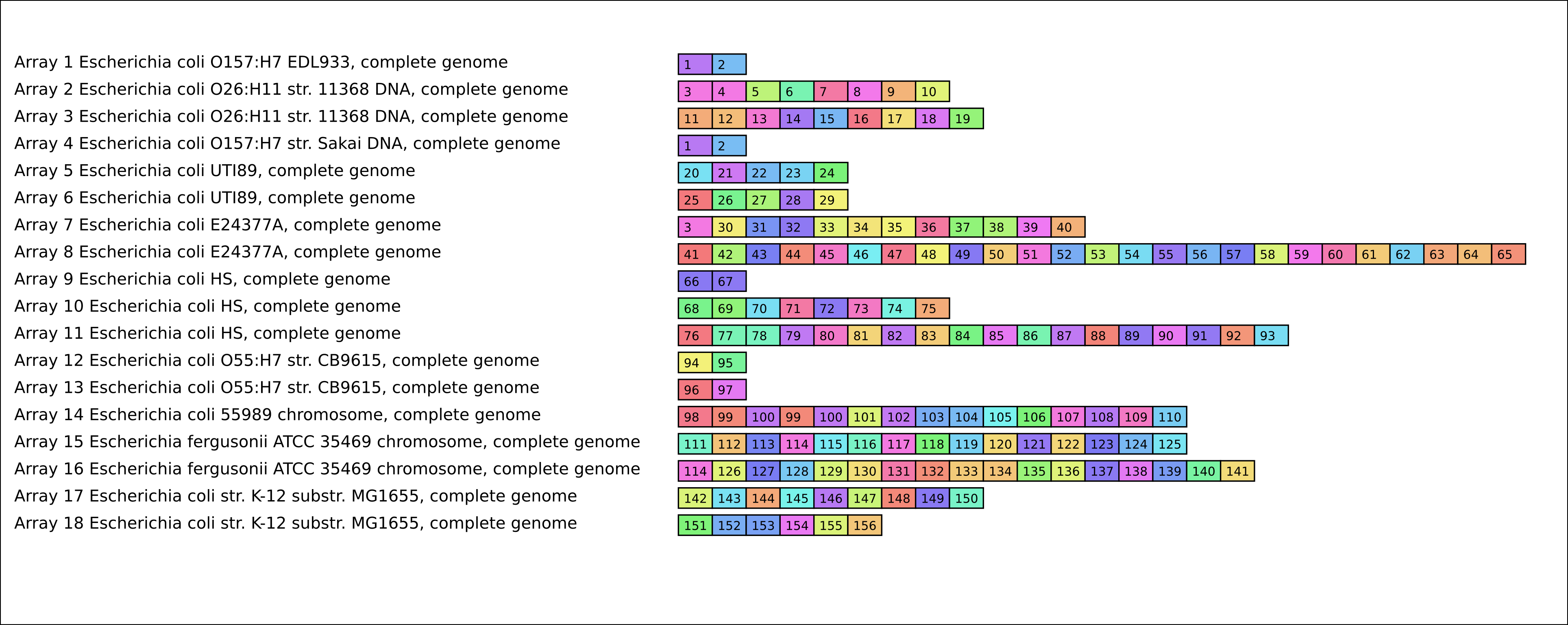
- <outputname> (text report from binCrisp itself, this defines the CRISPR sequence attached to each ID number in the figure at the bottom.)
- <outputname>.txt (text report of output from pilercr)
- <outputname>.svg (SVG Figure with a matrix of presence and absence of unique CRISPR sequence.)
See USAGE i.e. "python binCrisp.py findcr -h" for optional parameters.
Worked Example
The runex directory has a test case to check if binCrisp is running correctly. Dependencies include:
- Python 2.7+
- pilercr must be installed and on PATH: http://www.drive5.com/pilercr/
- svgwrite: https://pypi.python.org/pypi/svgwrite
binCrisp-Example.py is a quick example test of the binCrisp script. It requires reference genes and remote file list. (Default: fas-loc in runex dir). Just run:
'python binCrisp-Example.py -v'
This script runs an example case for the binCrisp script, based off 11 E. coli genomes (& E. fergusonii) downloaded from a remote server (listed in fas-loc). This script should be run from the runex folder in the parent binCrisp dir. This script will check Dependencies, format input files, and run binCrisp.py
binCrisp output will include:
- example (text report from binCrisp itself, this defines the CRISPR sequence attached to each ID number in the figure at the bottom.)
- example.txt (text report of output from pilercr)
- example.svg (SVG Figure with a matrix of presence and absence of unique CRISPR sequence.) [Shown below]
Total Runtime: ~3 minutes.
Licence
Nabil-Fareed Alikhan <n.alikhan@uq.edu.au>. (C) 2013.
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see <http://www.gnu.org/licenses/>.