Using persistent homology and multidimensional scaling on Wasserstein distance matrix
- S x C x N x N, where S = #subjects, C = #cohorts, N = #ROIs
- Mat file
- Size: 316 x 3 x 114 x 114
- Three cohorts: mx645, mx1400, std2500
- Removed NaN values by removing column 24 and row 24 from each 114 x 114 matrix
- Used correlation coefficients on transposed matrix and then applied square root on the 1 - squared distance
- 316 x 3 files, 3 files for each subject
- Each file contains 113 x 113 matrix
- Example:
subject_1_mx645.txt,subject_1_mx1400.txt,subject_1_std2500.txt
- Computed 0-dimensional persistent homology (PH) for all three cohorts of each subjects
- Generated 0-dimensional barcodes from calculated PH values with maximum value of 1
- To use persistent homology features from Gudhi library set
manual=Falseinget_barcodes_single_subjectmethod in distance_calculation.py. Otherwise, setmanual=Truefor raw calculation of persistent homology and 0-dimensional barcodes.
- Computed 1-Wasserstein Distance (WD) between cohorts for each subjects from the 0-dimensional barcodes
- For each subject, computed WD on the 0-dimensional barcodes:
- WD(mx645 - mx1400)
- WD(mx1400 - std2500)
- WD(std2500 - mx645)
- Generated 1 JSON file with 316 arrays, each array contains 3 values
- Generated file: distances_between_cohorts_ws.json
- Computed 0-dimensional persistent homology (PH) for all three cohorts of each subjects
- Generated 0-dimensional barcodes from calculated PH values with maximum value of 1
- To use persistent homology features from Gudhi library set
manual=Falseinget_barcodes_single_subjectmethod in distance_calculation.py. Otherwise, setmanual=Truefor raw calculation of persistent homology and 0-dimensional barcodes.
- Computed 1-Wasserstein Distance (WD) matrix within a cohort separately
- For each cohort, computed WD distance matrix on the 0-dimensional barcodes:
- WD_matrix(mx645): 316 x 316
- WD_matrix(mx1400): 316 x 316
- WD_matrix(std2500): 316 x 316
- Generated 3 JSON files each with 316 x 316 matrix
- Generated files:
- Applied classical metric Multidimensional scaling (MDS) with precomputed distance (1-Wasserstein)
- Calculated MDS of 2 components for each 1-Wasserstein distance matrix
- Generated 3 JSON files each with 316 x 2 matrix
- Generated files:
- Applied Kmeans++ clustering by selecting the number of clusters
nusing Silhouette Coefficient.
- Calculate
p-valueusing ANOVA test on the [316 x 3] size Wasserstein distances between the cohorts - ANOVA test p-value: 0.133
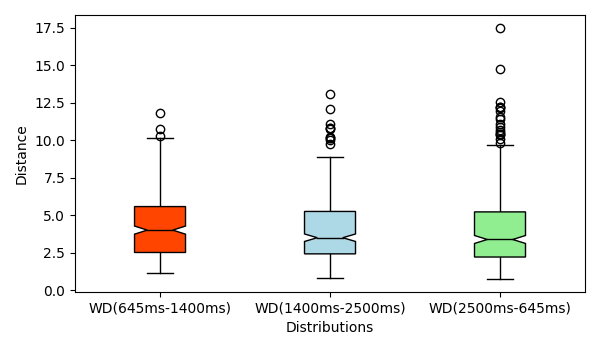
- Wasserstein distance for the following three pairs: (1) TR=645ms and TR=1400ms, (2) TR=1400ms and TR=2500ms and, (1) TR=2500ms and TR=645ms plotted using box plots: boxplots
- Plot WD distances between:
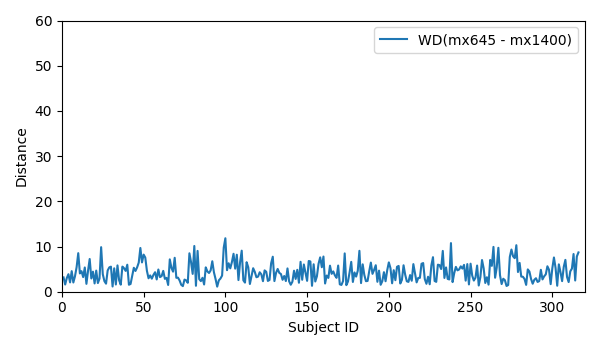
- WD for all 316 subjects for mx645 and mx1400: WD_mx645_mx1400
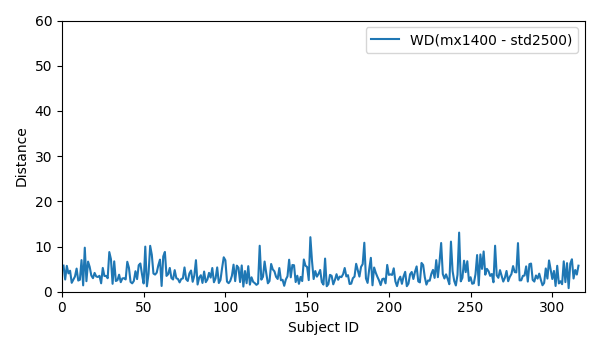
- WD for all 316 subjects for mx1400 and std2500: WD_mx1400_std2500
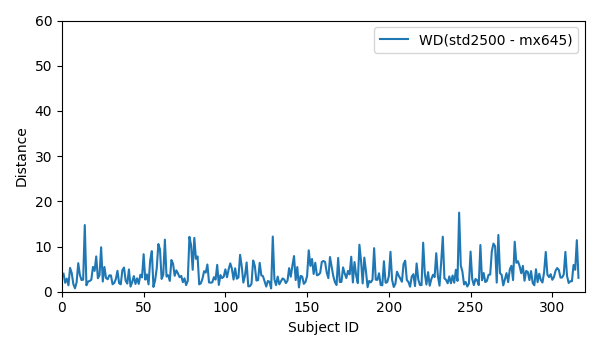
- WD for all 316 subjects for std2500 and mx645: WD_std2500_mx645
- Plot MDS value for all three cohorts: mds graph
- Clustering on the MDS results
- Wasserstein distance:
- Single figure: clustering_ws
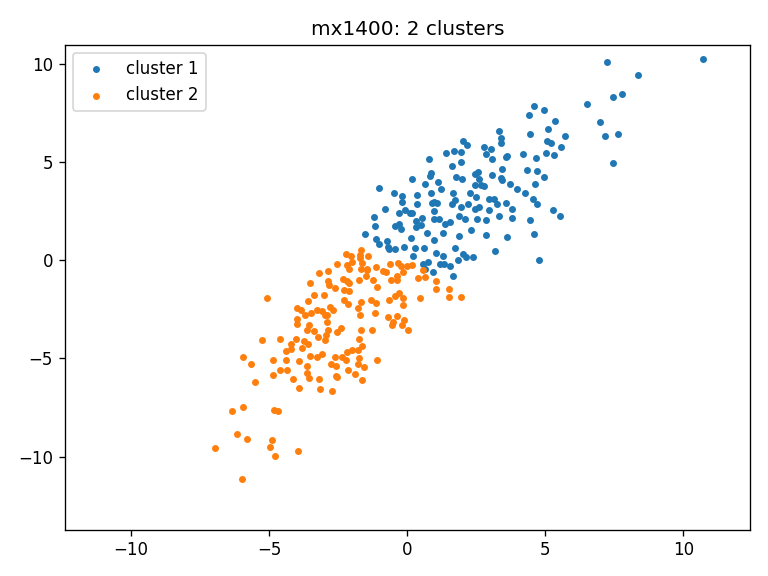
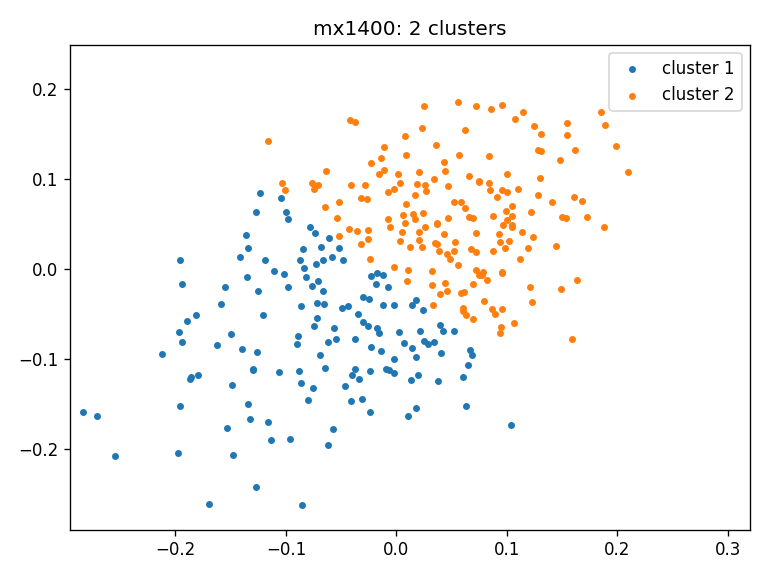
- mx1400: clusters_mx1400_ws
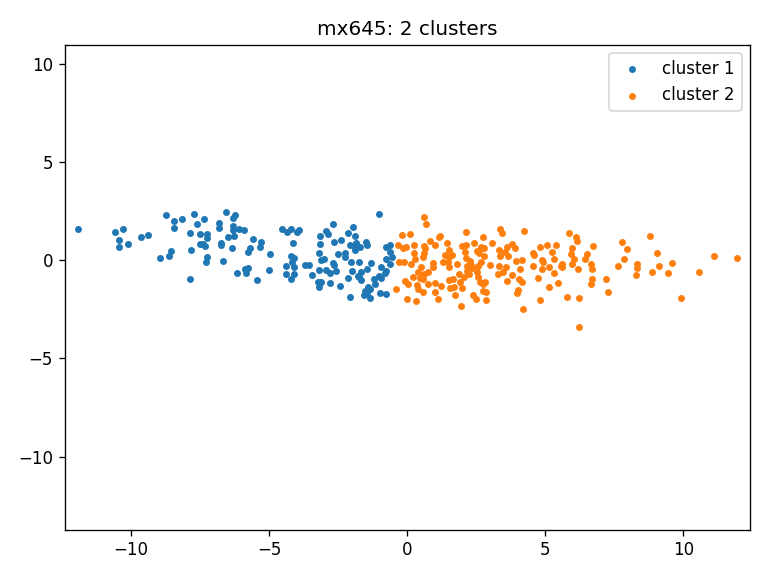
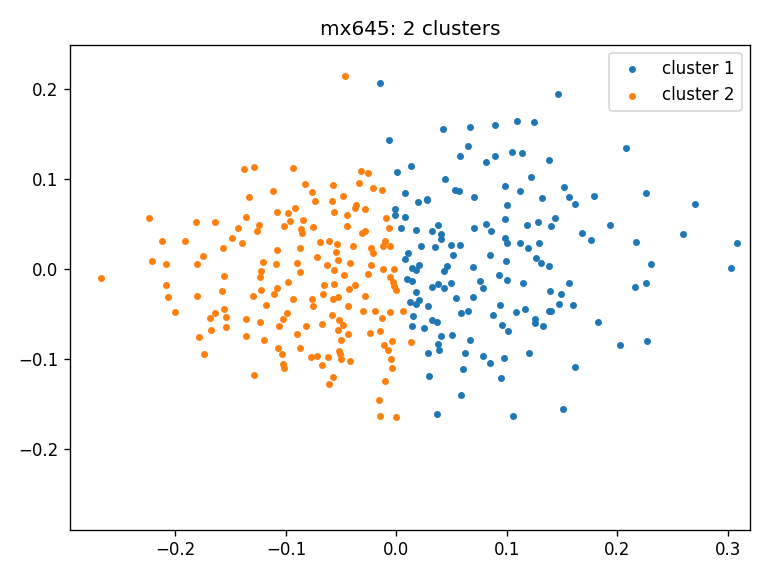
- mx645: clusters_mx645_ws
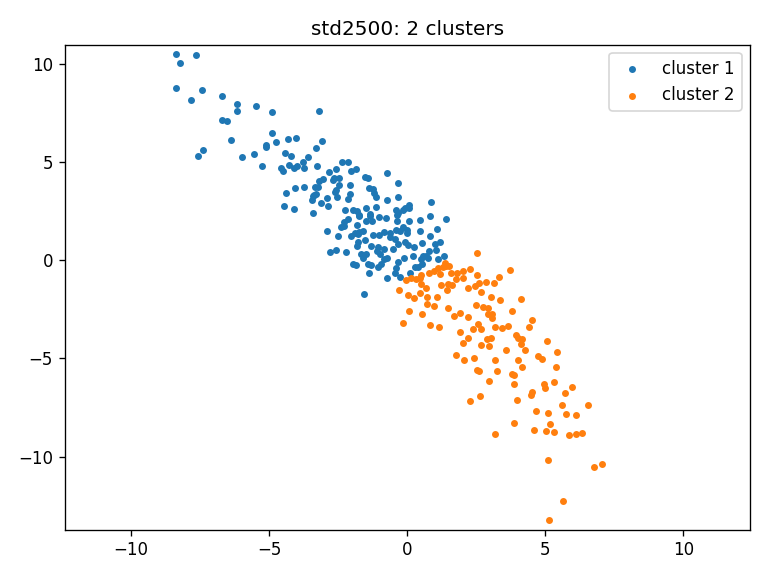
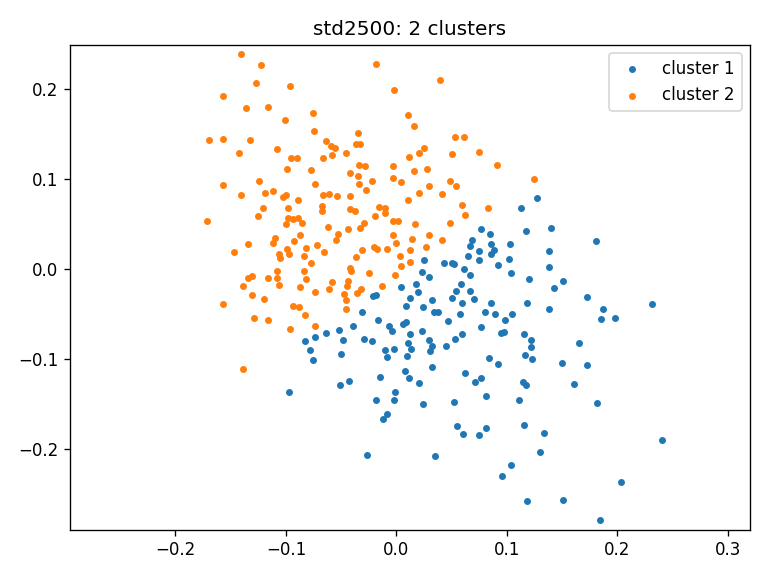
- std2500: clusters_std2500_ws
- Wasserstein distance:
- Python 3
- Clone the repository.
- Open a terminal / powershell in the cloned repository.
- Create a virtual environment and activate it. If you are using Linux / Mac:
python3 -m venv venv
source venv/bin/activate
Create and activate venv in Windows (Tested in Windows 10):
python -m venv venv
Set-ExecutionPolicy -ExecutionPolicy RemoteSigned -Scope CurrentUser
.\venv\Scripts\Activate.ps1
After activating venv, the terminal / powershell will have (venv) added to
the prompt.
- Check
pipversion:
pip --version
It should point to the pip in the activated venv.
- Install required packages:
pip install -r requirements.txt
- Calculate distance between cohorts and MDS within a cohort using WD:
python distance_calculation.py --method ws --start 1 --end 316 --distance y --mds y --data_dir full_data_linear --output_dir output_linear
- Draw plots and ANOVA test:
python statistical_calculation_linear.py --output_dir output_linear
- Generate clusters on the MDS data:
python cluster_calculation.py --output_dir output_linear
- Generate distance and MDS:
python distance_calculation.py --method ws --start 1 --end 316 --distance y --mds y --data_dir full_data_linear --output_dir output_linear- Running statistical analysis on the generated file:
python statistical_calculation_linear.py --output_dir output_linear
T-values:
0.059044 0.459634 0.286013
P-values:
0.088131 0.518936 0.387180
ANOVA test p-value: 0.289941
Mean WD_MX645_MX1400: 4.304
Mean WD_MX1400_STD2500: 4.01
Mean WD_STD2500_MX645: 4.135
WD_MX645_MX1400: Distance: 2, number of subjects: 42, percentage: 13.29%
WD_MX645_MX1400: Distance: 5, number of subjects: 207, percentage: 65.51%
WD_MX645_MX1400: Distance> 10, number of subjects: 4, percentage: 1.27%
Method main executed in 128.7125 seconds
- T-values and p-values obtained by pairwise t-tests comparing the WDs between data cohorts. Since all p-values are greater than 0.05, the means of WD distributions for each cohort comparison are statistically similar.
| t-value | p-value | ||
|---|---|---|---|
| WD(P1, P2) | WD(P2, P3) | 0.059044 | 0.088131 |
| WD(P2, P3) | WD(P3, P1) | 0.459634 | 0.518936 |
| WD(P3, P1) | WD(P1, P2) | 0.286013 | 0.387180 |
- Wasserstein distance for the following three pairs: (1) TR=645ms and
TR=1400ms, (2) TR=1400ms and TR=2500ms and, (1) TR=2500ms and TR=645ms
plotted using box plots:
- WD for all 316 subjects for mx645 and mx1400:
- WD for all 316 subjects for mx1400 and std2500:
- WD for all 316 subjects for std2500 and mx645:
- Clustering result for all three cohorts using Wasserstein distance:
- distances_between_cohorts_ws.json
- distance_matrix_mx645_ws.json
- distance_matrix_mx1400_ws.json
- distance_matrix_std2500_ws.json
- mds_mx645_ws.json
- mds_mx1400_ws.json
- mds_std2500_ws.json
- clustering_ws.json
python cluster_calculation.py --output_dir output_linear
Number of clusters in 3 cohorts: [2, 2, 2]
output_linear:
Cluster group: 000: #match: 24
Cluster group: 001: #match: 7
Cluster group: 010: #match: 26
Cluster group: 011: #match: 83
Cluster group: 100: #match: 115
Cluster group: 101: #match: 12
Cluster group: 110: #match: 20
Cluster group: 111: #match: 29
Max + reverse: 115 + 83 = 198
645-1400 : 236
1400-2500 : 251
2500-645 : 225
Adjacency matrix:
output_linear:
Rows X Columns: [645 clusters, 1400 clusters, 2500 clusters]
140 0 31 109 50 90
0 176 127 49 135 41
31 127 158 0 139 19
109 49 0 158 46 112
50 135 139 46 185 0
90 41 19 112 0 131 - Clustering result for full data for all three cohorts using Wasserstein distance:
python statistical_calculation_linear.py --output_dir output_linear
T-values:
0.059044 0.459634 0.286013
P-values:
0.088131 0.518936 0.387180
ANOVA test p-value: 0.289941- T-values and p-values obtained by pairwise t-tests comparing the WDs between data cohorts. Since all p-values are larger than 0.05, the means of WD distributions for each cohort comparison are statistically similar.
| t-value | p-value | ||
|---|---|---|---|
| WD(P1, P2) | WD(P2, P3) | 0.059044 | 0.088131 |
| WD(P2, P3) | WD(P3, P1) | 0.459634 | 0.518936 |
| WD(P3, P1) | WD(P1, P2) | 0.286013 | 0.387180 |
python cluster_calculation.py --output_dir output_random
Number of clusters in 3 cohorts: [2, 2, 2]
output_random:
Cluster group: 000: #match: 35
Cluster group: 001: #match: 38
Cluster group: 010: #match: 34
Cluster group: 011: #match: 43
Cluster group: 100: #match: 36
Cluster group: 101: #match: 32
Cluster group: 110: #match: 42
Cluster group: 111: #match: 56
Max + reverse: 56 + 35 = 91
Adjacency matrix:
output_random:
Rows X Columns: [645 clusters, 1400 clusters, 2500 clusters]
150 0 73 77 69 81
0 166 68 98 78 88
73 68 141 0 71 70
77 98 0 175 76 99
69 78 71 76 147 0
81 88 70 99 0 169 - Clustering result for random data for all three cohorts using Wasserstein distance:
Mean value of (Max + Reverse): 84.06122448979592
Standard deviation value of (Max + Reverse): 5.738786759358441- Within cohort: clustering
- Across cohorts: statistical analysis
- Original dataset: timeseries.Yeo2011.mm316.mat
- Total negative in correlation coefficient: 1234732 from all_positive_linear.m
- Total positive in correlation coefficient: 10870280 from all_negative_linear.m
- 3 out of 148 random dataset returns cluster
2, 2, 4, 1 returns4, 2, 2, and 144 returns2, 2, 2.
- non-TDA experiments for within cohort and comparison across cohort
- nonTDA on random for second pipeline
- create two matrices one for positive values and one for negative values and apply the distance function on them. Since, this will be a lot of experiments, if we do this for everything, let us just start by doing with only pipeline 1 (box plots, p/t-value tests). the original mat file which we normalized using matlab. 113 x 113 with all positive (padded by 0) and 113 x 113 with all negative (padded by 0).