Full documentation: https://psy-fer.github.io/SquiggleKitDocs/
publication: SquiggleKit: A toolkit for manipulating nanopore signal data
Pre-print: SquiggleKit: A toolkit for manipulating nanopore signal data
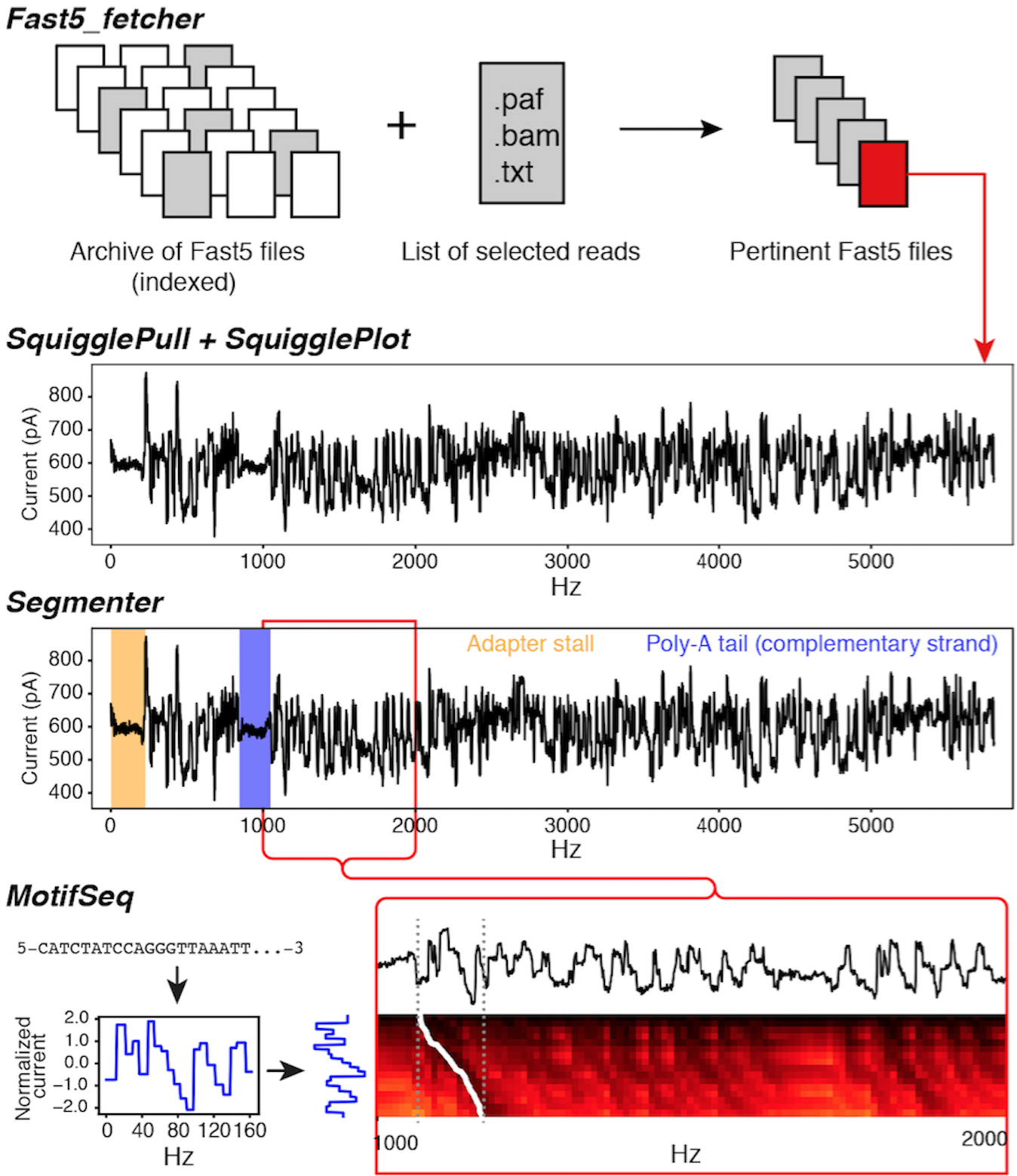
Fast5_fetcher: merge single files into multi-fast5 files
SquigglePull: python3, read from multi-fast5
SquigglePlot: python3, read from multi-fast5, image size args, arg clean-up
Segmenter: dynamic file formats and more stability
MotifSeq: Improved background modelling, custom modelling, RNA specific tools, custom alignment methods
| Tool | Category | Description |
|---|---|---|
| Fast5_fetcher | File management |
Fetches fast5 files given a filtered input list |
| SquigglePull | Signal extraction |
Extracts event or raw signal from data files |
| SquigglePlot | Signal visualisation |
Visualisation tool for signal data |
| Segmenter | Signal analysis |
Finds adapter stall, and homopolymer regions |
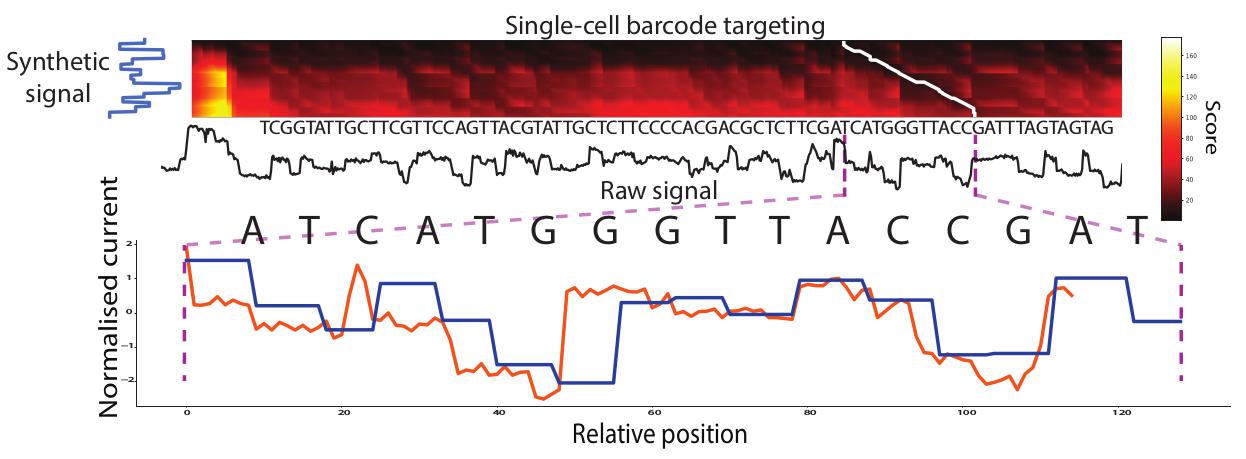
| MotifSeq | Signal analysis |
Finds nucleotide sequence motifs in signal, i.e.“Ctrl+F” |
Following a self imposed guideline, most things written to handle nanopore data or bioinformatics in general, will use as little 3rd party libraries as possible, aiming for only core libraries, or have all included files in the package.
In the case of fast5_fetcher.py and batch_tater.py, only core python libraries are used. So as long as Python 2.7+ is present, everything should work with no extra steps.
There is one catch. Everything is written primarily for use with Linux. Due to MacOS running on Unix, so long as the GNU tools are installed (see below), there should be minimal issues running it. Windows however may require more massaging. The Windows-Subsystem-Linux must be installed. Follow the instructions here to do this.
SquiggleKit tools were not made to be executable to allow for use with varying python environments on various operating systems. To make them executable, add #! paths, such as #!/usr/bin/env python2.7 as the first line of each of the files, then add the SquiggleKit directory to the PATH variable in ~/.bashrc, export PATH="$HOME/path/to/SquiggleKit:$PATH"
git clone https://github.com/Psy-Fer/SquiggleKit.git
Use pip for python 2 and pip3 for python 3. User environments may vary.
for
fast5_fetcher.py, SquigglePull.py, SquigglePlot.py segmenter.py:
- numpy
- matplotlib
- h5py
- sklearn
- ont_fast5_api
- pyslow5
pip install numpy h5py sklearn matplotlib
# do this after to get around an annoying version check bug
pip install pyslow5
for MotifSeq.py:
- all of the above
- scipy
- scrappie
- mlpy 3.5.0 (only use pip3 in python 3 - see below)
pip install scipy scrappie
- Download the Files
- Install Instructions
pip3 install machine-learning-py
If your fast5 files are compressed with ONT VBZ, then you will need to install their VBZ plugin for hdf5/h5py
https://github.com/nanoporetech/vbz_compression/releases
This will fix errors like this:
OSError: Can't read data (can't open directory: /home/jamfer/squigglekit_env/lib/hdf5/plugin)
extract_fast5_all():failed to read readID: read_6c3c3be4-e5b0-89aa-b0a0-a1c0167ce136 Traceback (most recent call last):
File "SquigglePull.py", line 211, in extract_f5_all
for col in hdf[read]['Raw/Signal'][()]:
File "h5py/_objects.pyx", line 54, in h5py._objects.with_phil.wrapper
File "h5py/_objects.pyx", line 55, in h5py._objects.with_phil.wrapper
If using MacOS, and NOT using homebrew, install it here:
homebrew installation instructions
then install gnu-tar with:
brew install gnu-tar
How the index is built depends on which file structure you are using. It will work with both tarred and un-tarred file structures. Tarred is preferred. (zip and other archive methods are being investigated)
for file in $(pwd)/reads/*/*;do echo $file; done >> name.index
gzip name.indexfor file in $(pwd)/reads.tar; do echo $file; tar -tf $file; done >> name.index
gzip name.indexfor file in $(pwd)/fast5/*fast5.tar; do echo $file; tar -tf $file; done >> name.indexIf you have multiple experiments, then cat them all together and gzip.
for file in ./*.index; do cat $file; done >> ../all.name.index
gzip all.name.indexusing a filtered paf file as input:
python fast5_fetcher.py -p my.paf -s sequencing_summary.txt.gz -i name.index.gz -o ./fast5All raw data:
python SquigglePull.py -rv -p ~/data/test/reads/1/ -f all > data.tsv
Positional event data:
python SquigglePull.py -ev -p ./test/ -t 50,150 -f pos1 > data.tsv
Plot individual fast5 file:
python SquigglePlot.py -i ~/data/test.fast5
Plot files in path
python SquigglePlot.py -p ~/data/ --plot_colour -g
Plot first 2000 data points of each read from signal file and save at 300dpi pdf:
python SquigglePlot.py -s signals.tsv.gz --plot_colour teal -n 2000 --dpi 300 --no_show o--save test.pdf --save_path ./test/plots/
Identify any segments in folder and visualise each one
Use f to full screen a plot, and ctrl+w to close a plot and move to the next one.
python segmenter.py -p ./test/ -v
Stall identification
python segmenter.py -s signals.tsv.gz -ku -j 100 > signals_stall_segments.tsv
Find kmer motif:
fasta format for model:
my_kmer.fa
>my_kmer_name
ATCGATCGCTATGCTAGCATTACG
find the best match to that k-mer in the signal:
python MotifSeq.py -s signals.tsv -i my_kmer.fa > signals_kmer.tsv
k-mer length should not really be below 12nt, below this things get hairy based on modelling
The p-values and hit probabilities provided are based on loose modelling of negative background scores for a number of k-mers. It is currently only modelled on R9.4 model, not R10 or RNA.
I would like to thank the members of my lab, Shaun Carswell, Kirston Barton, Hasindu Gamaarachchi, Kai Martin, Tansel Ersavas, Brent Bevear, Jillian Hammond, and Martin Smith, from the Genomic Technologies team from the Garvan Institute for their feedback on the development of these tools.