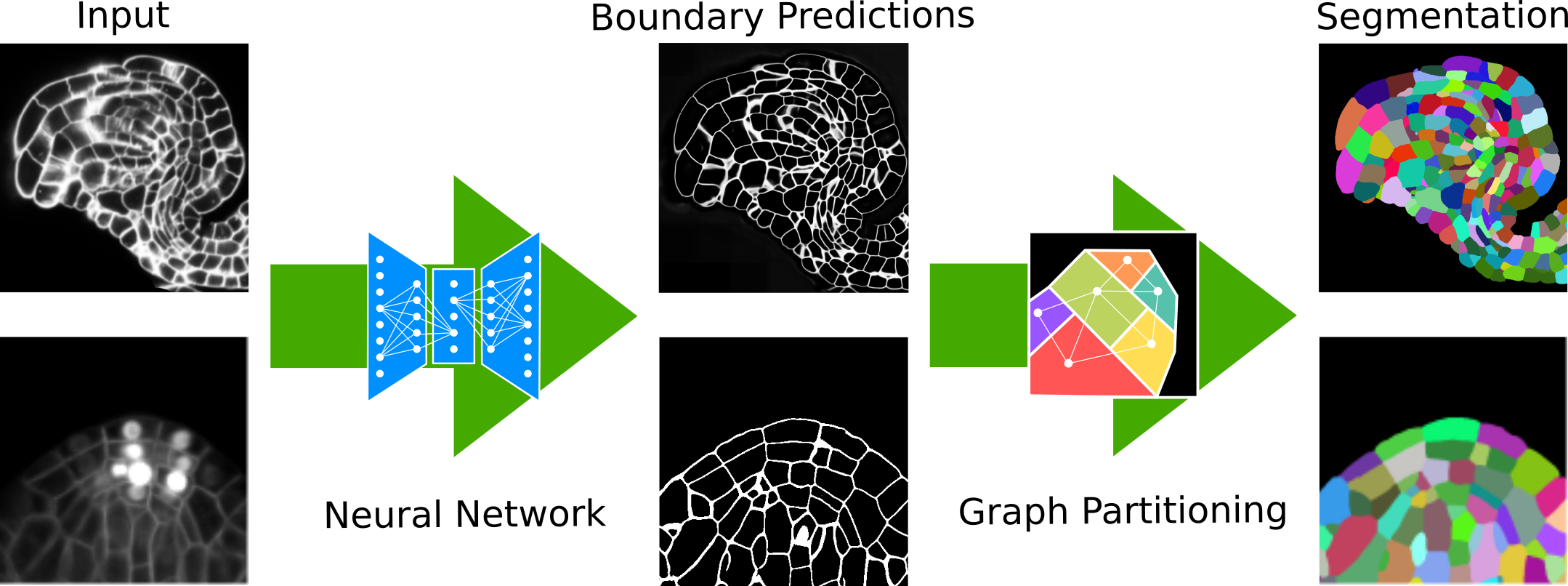
PlantSeg is a tool for cell instance aware segmentation in densely packed 3D volumetric images. The pipeline uses a two stages segmentation strategy (Neural Network + Segmentation). The pipeline is tuned for plant cell tissue acquired with confocal and light sheet microscopy. Pre-trained models are provided.
- Getting Started
- Install PlantSeg
- Repository Index
- Datasets
- Pre-trained networks
- Training on New Data
- Citation
For detailed usage checkout our documentation 📖.
| Documentation | Napari GUI | Legacy GUI | Command Line |
|---|---|---|---|
 |
 |
 |
 |
Please go to the documentation for more detailed instructions. In short, we recommend using mamba to install PlantSeg, which is currently supported on Linux and Windows.
-
GPU version, CUDA=12.x
mamba create -n plant-seg -c pytorch -c nvidia -c conda-forge pytorch pytorch-cuda=12.1 pyqt plant-seg
-
GPU version, CUDA=11.x
mamba create -n plant-seg -c pytorch -c nvidia -c conda-forge pytorch pytorch-cuda=11.8 pyqt plant-seg
-
CPU version
mamba create -n plant-seg -c pytorch -c nvidia -c conda-forge pytorch cpuonly pyqt plant-seg
The above command will create new conda environment plant-seg together with all required dependencies.
The PlantSeg repository is organised as follows:
- plantseg: Contains the source code of PlantSeg.
- conda-reicpe: Contains all necessary code and configuration to create the anaconda package.
- Documentation-GUI: Contains a more in-depth documentation of PlantSeg functionality.
- evaluation: Contains all script required to reproduce the quantitative evaluation in Wolny et al..
- examples: Contains the files required to test PlantSeg.
- tests: Contains automated tests that ensures the PlantSeg functionality are not compromised during an update.
We publicly release the datasets used for training the networks which available as part of the PlantSeg package. Please refer to our publication for more details about the datasets:
- Arabidopsis thaliana ovules dataset (raw confocal images + ground truth labels)
- Arabidopsis thaliana lateral root (raw light sheet images + ground truth labels)
Both datasets can be downloaded from our OSF project
The following pre-trained networks are provided with PlantSeg package out-of-the box and can be specified in the config file or chosen in the GUI.
generic_confocal_3D_unet- alias forconfocal_3D_unet_ovules_ds2xsee belowgeneric_light_sheet_3D_unet- alias forlightsheet_3D_unet_root_ds1xsee belowconfocal_3D_unet_ovules_ds1x- a variant of 3D U-Net trained on confocal images of Arabidopsis ovules on original resolution, voxel size: (0.235x0.075x0.075 µm^3) (ZYX) with BCEDiceLossconfocal_3D_unet_ovules_ds2x- a variant of 3D U-Net trained on confocal images of Arabidopsis ovules on 1/2 resolution, voxel size: (0.235x0.150x0.150 µm^3) (ZYX) with BCEDiceLossconfocal_3D_unet_ovules_ds3x- a variant of 3D U-Net trained on confocal images of Arabidopsis ovules on 1/3 resolution, voxel size: (0.235x0.225x0.225 µm^3) (ZYX) with BCEDiceLossconfocal_2D_unet_ovules_ds2x- a variant of 2D U-Net trained on confocal images of Arabidopsis ovules. Training the 2D U-Net is done on the Z-slices (1/2 resolution, pixel size: 0.150x0.150 µm^3) with BCEDiceLossconfocal_3D_unet_ovules_nuclei_ds1x- a variant of 3D U-Net trained on confocal images of Arabidopsis ovules nuclei stain on original resolution, voxel size: (0.35x0.1x0.1 µm^3) (ZYX) with BCEDiceLosslightsheet_3D_unet_root_ds1x- a variant of 3D U-Net trained on light-sheet images of Arabidopsis lateral root on original resolution, voxel size: (0.25x0.1625x0.1625 µm^3) (ZYX) with BCEDiceLosslightsheet_3D_unet_root_ds2x- a variant of 3D U-Net trained on light-sheet images of Arabidopsis lateral root on 1/2 resolution, voxel size: (0.25x0.325x0.325 µm^3) (ZYX) with BCEDiceLosslightsheet_3D_unet_root_ds3x- a variant of 3D U-Net trained on light-sheet images of Arabidopsis lateral root on 1/3 resolution, voxel size: (0.25x0.4875x0.4875 µm^3) (ZYX) with BCEDiceLosslightsheet_2D_unet_root_ds1x- a variant of 2D U-Net trained on light-sheet images of Arabidopsis lateral root. Training the 2D U-Net is done on the Z-slices (pixel size: 0.1625x0.1625 µm^3) with BCEDiceLosslightsheet_3D_unet_root_nuclei_ds1x- a variant of 3D U-Net trained on light-sheet images Arabidopsis lateral root nuclei on original resolution, voxel size: (0.25x0.1625x0.1625 µm^3) (ZYX) with BCEDiceLosslightsheet_2D_unet_root_nuclei_ds1x- a variant of 2D U-Net trained on light-sheet images Arabidopsis lateral root nuclei. Training the 2D U-Net is done on the Z-slices (pixel size: 0.1625x0.1625 µm^3) with BCEDiceLoss.confocal_3D_unet_sa_meristem_cells- a variant of 3D U-Net trained on confocal images of shoot apical meristem dataset from: Jonsson, H., Willis, L., & Refahi, Y. (2017). Research data supporting Cell size and growth regulation in the Arabidopsis thaliana apical stem cell niche. https://doi.org/10.17863/CAM.7793. voxel size: (0.25x0.25x0.25 µm^3) (ZYX)confocal_2D_unet_sa_meristem_cells- a variant of 2D U-Net trained on confocal images of shoot apical meristem dataset from: Jonsson, H., Willis, L., & Refahi, Y. (2017). Research data supporting Cell size and growth regulation in the Arabidopsis thaliana apical stem cell niche. https://doi.org/10.17863/CAM.7793. pixel size: (25x0.25 µm^3) (YX)lightsheet_3D_unet_mouse_embryo_cells- A variant of 3D U-Net trained to predict the cell boundaries in live light-sheet images of ex-vivo developing mouse embryo. Voxel size: (0.2×0.2×1 µm^3) (XYZ)confocal_3D_unet_mouse_embryo_nuclei- A variant of 3D U-Net trained to predict the cell boundaries in live light-sheet images of ex-vivo developing mouse embryo. Voxel size: (0.2×0.2×1 µm^3) (XYZ)
Selecting a given network name (either in the config file or GUI) will download the network into the ~/.plantseg_models
directory.
Detailed description of network training can be found in our paper.
The PlantSeg home directory can be configured with the PLANTSEG_HOME environment variable.
export PLANTSEG_HOME="/path/to/plantseg/home"For training new models we rely on the pytorch-3dunet.
A similar configuration file can be used for training on new data and all the instructions can be found in the repo.
When the network is trained it is enough to create ~/.plantseg_models/MY_MODEL_NAME directory
and copy the following files into it:
- configuration file used for training:
config_train.yml - snapshot of the best model across training:
best_checkpoint.pytorch - snapshot of the last model saved during training:
last_checkpoint.pytorch
The later two files are automatically generated during training and contain all neural networks parameters.
Now you can simply use your model for prediction by setting the model_name key to MY_MODEL_NAME.
If you want your model to be part of the open-source model zoo provided with this package, please contact us.
@article{wolny2020accurate,
title={Accurate and versatile 3D segmentation of plant tissues at cellular resolution},
author={Wolny, Adrian and Cerrone, Lorenzo and Vijayan, Athul and Tofanelli, Rachele and Barro, Amaya Vilches and Louveaux, Marion and Wenzl, Christian and Strauss, S{\"o}ren and Wilson-S{\'a}nchez, David and Lymbouridou, Rena and others},
journal={Elife},
volume={9},
pages={e57613},
year={2020},
publisher={eLife Sciences Publications Limited}
}