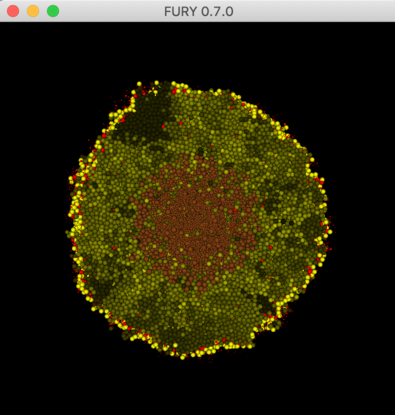
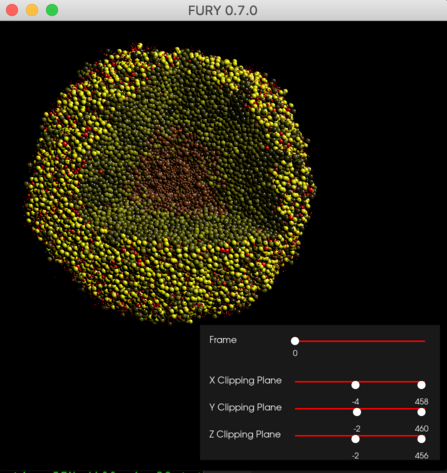
Use Fury to visualize output from a PhysiCell 3-D model.
Assuming you have installed the Anaconda Python, you can install Fury using its pip command:
pip install furyAfter it finishes installing, you should be able to run this script (which uses files in the /data directory).
python tumor_vis.pyWhen the Fury window appears, you should be able to interact with the data using your mouse or trackpad:
- click (mouse: button 1), drag to rotate
- shift- click (mouse: button 2), drag to pan
- shift-control click (mouse: button 3), drag to zoom
Compare the visualization with this video at about 15 secs from the start.
This next script provides sliders for slicing planes to let you "cut" into the 3-D tumor, and also a slider to step through time steps of data.
python tumor_vis_sliders.py(after rotating and panning the tumor)