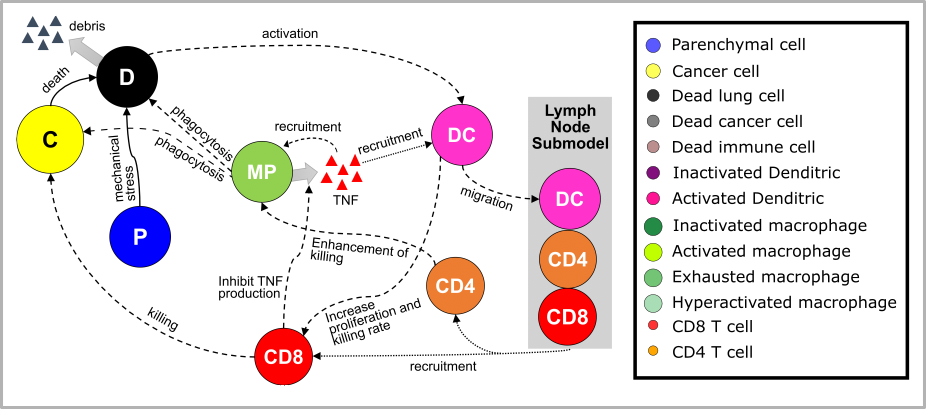
Building off of recent COVID-19 modeling (https://www.biorxiv.org/content/10.1101/2020.04.02.019075v4), we developed an agent-based model of immune surveillance in micrometastases. The model includes cancer cells, parenchymal cells, key immune cell types, and cytokine-mediated and mechanical interactions. Immune cells traffic to and from the lymphatic system to drive an expanding immune response.
This model is part of the pilot project for building a Cancer patient digital twins (CPDTs) framework.
To compile and run PhysiCell for this model:
# your compiler needs to support OpenMP
$ make
$ ./micromets_immuno
Requirements: our analysis have been performed using Python 3.9.18. The following modules are required to run the jupyter notebook:
- Matplotlib 3.8.0 (https://matplotlib.org/)
- SciPy 1.11.2 (https://scipy.org/)
- pandas 1.5.3 (https://pandas.pydata.org/)
- wget 3.2 (https://pypi.org/project/wget/)
- torchvision 0.15.2 (https://pytorch.org/vision/stable/index.html)
- fitter 1.5.2 (https://fitter.readthedocs.io/en/latest/)
- matplotlib_venn 0.11.9 (https://pypi.org/project/matplotlib-venn/)
- seaborn 0.12.2 (https://seaborn.pydata.org/)
- statannot 0.2.3 (https://pypi.org/project/statannot/)
If you are using Anaconda, you can create the conda environment manually:
conda create -n micromets python=3.9
conda activate micromets
pip install jupyter pandas==1.5.3 matplotlib==3.8.0 wget==3.2 torchvision==0.15.2 scipy==1.11.2 fitter==1.5.2 matplotlib_venn==0.11.9 seaborn==0.12.2 statannot==0.2.3
or you can create from the micromets.yml file:
conda env create -f micromets.yml
Attention: In this dataset, when interpreting cell annotations, replace 'lung cells' with 'parenchyma cells'. It's important to note that this modification does not impact the analysis results. Originally, the dataset was based on an initial hypothesis suggesting a focus on lung tissue, but the current interpretation categorizes it as general epithelial tissue.