AmiraMesh 3D ASCII 2.0 parser
This repository has code and test files for AmiraMesh 3D ASCII 2.0 format for passing on branching and thickness information from reconstructed biological cells.
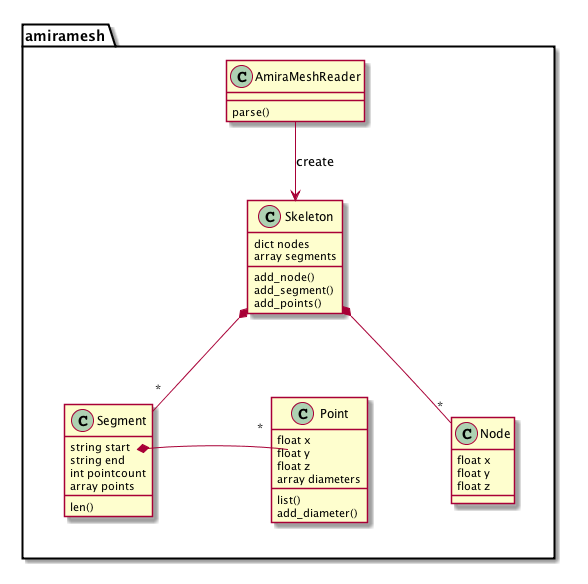
Code structure
A 3D skeleton structure in amiramesh consists of nodes in a graph and segments that join them. Points along the segments are given to show the path of the segment. Each point also knows about approximate diameter or two at the position.
Testing
The code has been tested in Python 2.7.6.
bin/read_ameramesh.py -v on sample datafiles should print out
the same numbers that are given in the datafile header.
Testing was done with nose and coverage to 100% coverage:
nosetests --with-coverage --cover-package amiramesh