Helao deploys Hierachical Experimental Laboratory Automation and Orchestration The idea behind this project is that we wanted to deploy active learning to different devices located in our laboratory and to others and even to many instruments running in parallel. To to this we utilized fastAPI, a web framework that allows the facile developement of APIs.
You may find the preprint on chemRxiv here: here
The hdf5 files created during a simulated active learning run both in parallel and sequential instrument operation can be found here
This repository is our developement branch as of the day of publication. If you seek to make changes please do so but this will need permission from some of the contributors.
Materials acceleration platforms (MAPs) operate on the paradigm of integrating combinatorial synthesis, high-throughput characterization, automatic analysis and machine learning. Within these MAPs one or multiple autonomous feedback loops aim to optimize materials for certain functional properties or generate new insights. Fundamentally, this necessitates accelerated, but foremost integrated, research actions. Herein, a web based asynchronous protocol to seamlessly integrate research tasks within a hierarchical experimental laboratory automation and orchestration (HELAO) framework is presented. We demonstrate the capability of orchestrating distributed research instruments that may incorporate data from experiments, simulations, and databases. HELAO offers interfacing laboratory hardware and software distributed across several computers and operating systems for executing an experiment, data analysis, provenance tracking, and autonomous planning. Research acceleration in terms of reduction of total experimental time is demonstrated to be close to 2x (in addition to the speedup of active learning of 5-10x depending on active learning metric) by deploying a centrally orchestrated fleet of instruments for a active learning. To the best of our knowledge, HELAO is the only laboratory automation framework with integrated data management capable of running closed loop optimization on multiple instruments and extreme modularity.
If you want to build autonomous labs that are spread around the globe use this framework - it might save you a lot of stress.
We have implemented a series of drivers and analysis techniques which you can easily reuse if you have the same or similar hardware. Currently implemented devices in the laboratories at KIT and Caltech are shown in the table below. Instruments build from this include scanning droplet cells, high-throughput spectrometers and a battery assembly robot (all to be published elsewhere). The extreme modularity allows to mix and match any of these devices by simply defining a sequence of events i.e. to build an integrated SDC and spectrometer or a sample echange robot no code changes are necessary. The currently implemented hardware is the following:
| Device Name | Type | Communication | Measures | Manufacturer | natively blocking |
|---|---|---|---|---|---|
| lang | Motion | .net API | position | Lang GmbH | no |
| galil | Motion, IO | TCP/IP | position | Galil Motion Control Inc. | no |
| owis | Motion | serial commands | position | Owis GmbH | no |
| mecademic | Motion | python TCP/IP API | position, state | Mecademic Ltd. | no |
| rail | Motion | TCP/IP | position | Jenny Science AG | no |
| autolab | Potentiostat | .net API | electrochemistry | Methrohm Autolab B.V. | yes |
| gamry | Potentiostat | .dll for serial communication | electrochemistry | Gamry Instruments Inc. | yes |
| arbin | Potentiostat | autohotkey | electrochemistry | Arbin Inc. | no |
| pump | pumping | serial commands | n.a. | CAT engineering GmbH | no |
| arcoptix | spectroscopy | .dll api | IR spectra | arcoptix S.A. | yes |
| ocean | spectroscopy Raman | python package | Raman spectra | ocean insights GmbH | yes |
| force | force sensing | serial commands | force | ME Meßsysteme GmbH | n/a |
| arduino | relays, I/O | python package | n.a. | arduino | no |
| kadi | data management | python package | n.a. | KIT | yes |
| aux | machine learning and analysis | python package | n.a. | n.a. | yes |
There are dummy drivers and dummy analysis "devices" indicating how you can your own.
HELAO is very lightweight and besides hardware drivers you just need a working python installation with fastAPI and starlette. If you wish to setup thing super easy from scratch just follow these steps:
- install miniconda[https://docs.conda.io/en/latest/miniconda.html], python 3 only
- clone git repository
- from repo directory, setup conda environment using
conda env create -f helao.yml
- galil and gamry server code current import from driver.*_simulate
- cd into server directory, execute start fastapi instances via
python galil_server.pyandpython gamry_server.py
helao.pyscript can validate server configuration parameters, launch a group of servers, and shutdown all servers beloning to a group- server groups may be defined as .py files in the
config/folder (seeconfig/world.pyas an example) - launch syntax:
python helao.py worldwill validate and launch servers with parameters defined inconfig/world.py, while also writing all monitored process IDs topids_world.pckin the root directory - exercise caution when running multiple server groups as there is currently no check for ports that are currently in-use between different config files
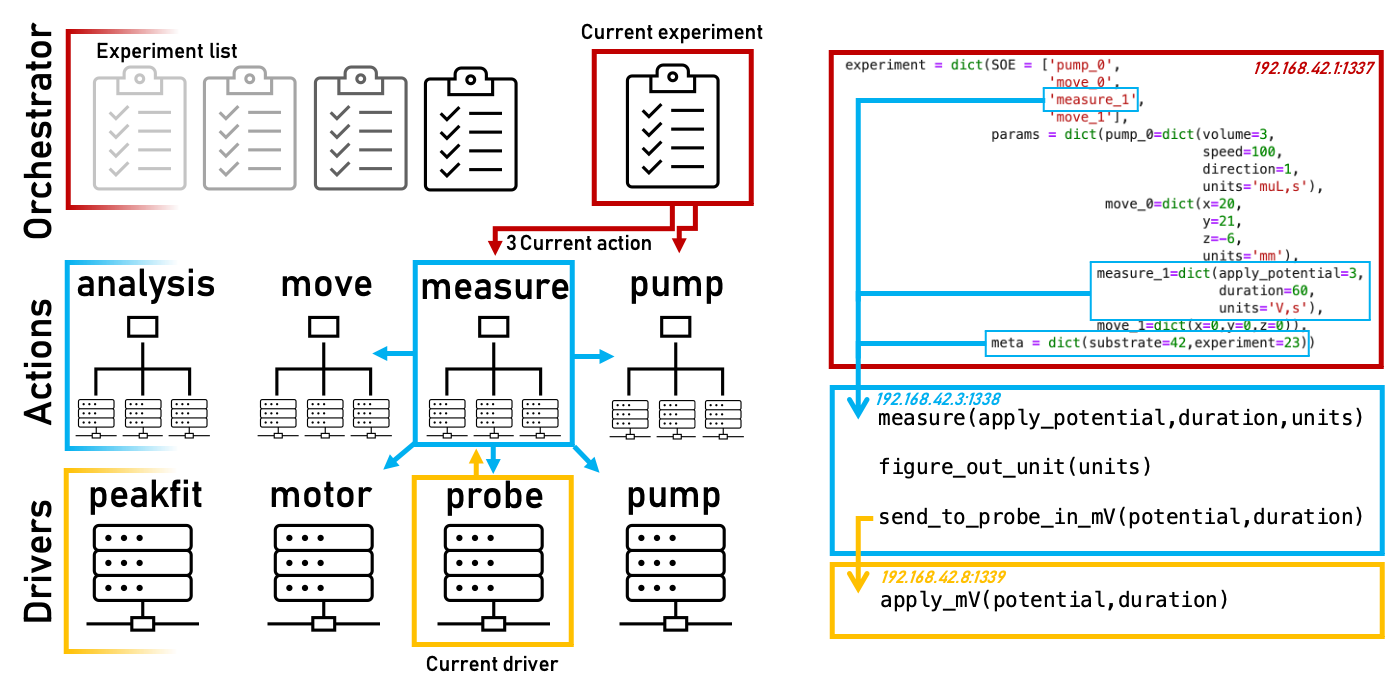
High level layout of HELAO where experiments are executed by sequentially calling actions which are high level wrappers for other actions or low level driver instructions. Communication goes hierarchically down from the orchestrator level to actions, which may communicate with one another, to the lowest level of drivers which may only communicate with the calling action. The orchestrator, actions and drivers are all exposing python class functions through a web interface allowing for a modular and distributed hosting of each item. Experiments are dictionaries containing a sequence of events (SOE) that outlines in which the actions are to be executed. All actions require parameters and are supplied with experiment level metadata. Metadata may be introduced at any level.
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 957189.