Codes for the 2023 ETH Zürich course Advanced Machine Learning.
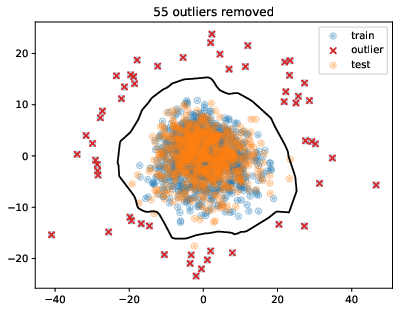
- Outlier removal
- Remove median and scale data according to interquartile range
- Imputation for missing values using k-Nearest Neighbors
- Principal component analysis, reduction to 2 components
- Isolation Forest Algorithm, with contamination of 4.5% to detect/remove outliers (55)
- Preprocessing + feature selection
- Standardize features by removing the mean and scaling to unit variance
- Imputation for missing values using k-Nearest Neighbors
- Remove features that have zero variance
- Select the 200 features that have the highest correlation with target
- Select features based on importance weights of Lasso regression (74)
- Model selection
- Stacked regression model consisting of
- Support Vector Regression
- Histogram-based Gradient Boosting Regression Tree
- Extra-trees regressor
- Multi-layer Perceptron regressor
- All hyperparameters are found/validated through 10-fold cross-validated grid search
- Stacked regression model consisting of
- feature extraction
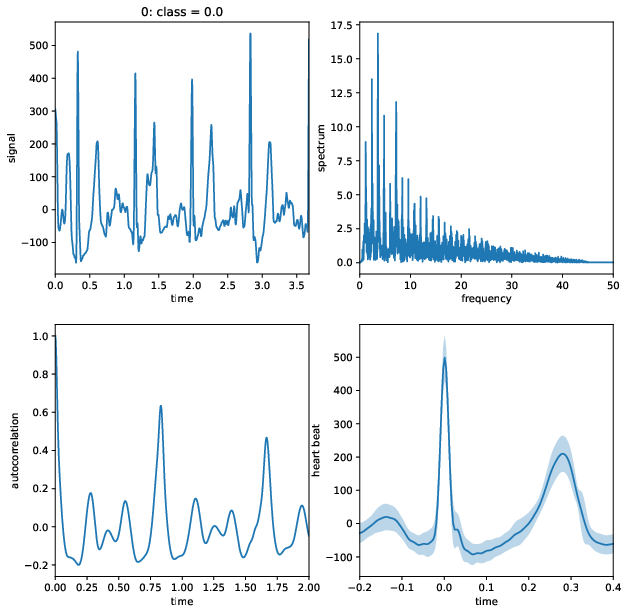
- use biosppy to extract raw features (cleaned signal, rpeaks, heart beats, heart rate)
- find S, Q, P, and T points using neurokit
- some of the signals are inverted, to combat this we add the inverse of all signals
- use binned FFT and autocorrelation of full spectrum
- compute various time intervals between R, S, Q, P, and T points (and their on/offsets)
- use mean, standard deviation, median, and variance of the features
- preprocessing
- because the dataset is imbalanced, we use random over sampling
- scale every feature to zero mean and unit variance
- training
- use HistGradientBoostingClassifier from sklearn
- optimize hyper parameters with RandomizedGridSearchCV
- preprocessing
- images are padded to be square and rescaled to 128x128 pixels
- move the frame axis of videos and labels to the front
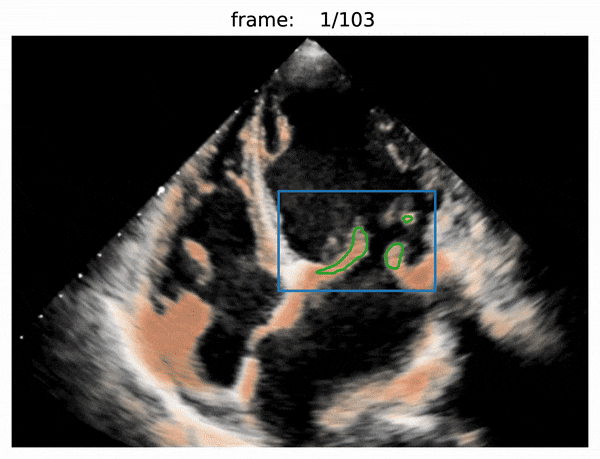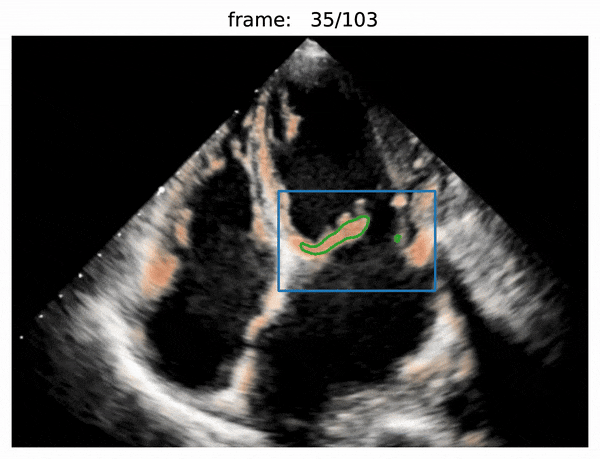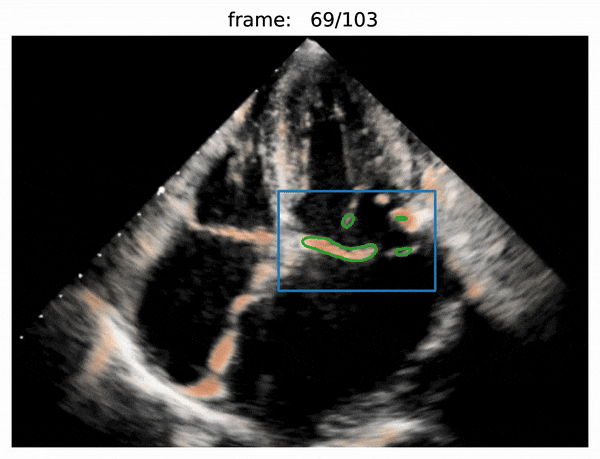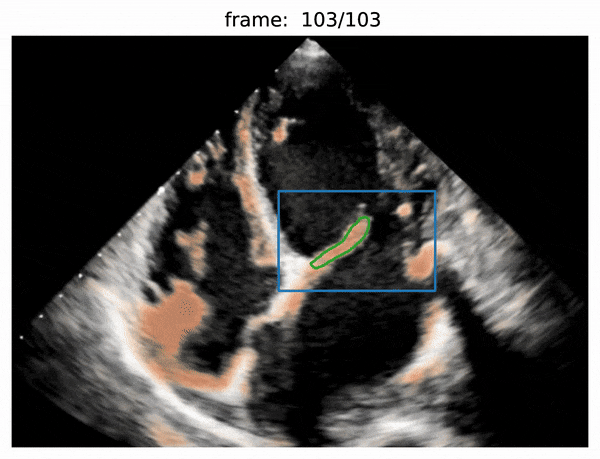
- box prediction
- use Attention R2U-Net to predict boxes
- use Adam optimizer and BCEWithLogitsLoss loss
- augment images by
- random erasing of image portion
- random affine transformation (rotate, translate, scale, shear)
- random perspective
- train first on amateur data, then on expert data
- split train/validation set over patients not frames
- train with batch size 16, until the best validation score does not decrease for 3 epochs
- average all boxes for one video, use threshold of 0.5 to create binary mask
- movement computation
- use robust non-negative matrix factorization to detect moving pixels in videos
- rank=2 and sparsity=0.1
- normalize videos and movement to be between 0 and 1
- valve segmentation
- use Attention R2U-Net to predict boxes
- use Adam optimizer and BCEWithLogitsLoss loss
- augment images by
- random erasing of image portion in the shape of a circle from a random point in the label
- mimics occlusion of valve in unlabeled frames
- random affine transformation (rotate, translate, scale, shear)
- random perspective
- random erasing of image portion in the shape of a circle from a random point in the label
- assemble inputs as 3 channel stacked tensors (frame, box, movement)
- train first on amateur data, then on expert data
- split train/validation set over patients not frames
- train with batch size 8, until the best validation score does not decrease for 5 epochs
- use threshold of 0.5 for each prediction
- postprocessing
- rescale to original size and remove padding
- move frame axis back to the rear