Code and figures for "Controlling the speed and trajectory of evolution with counterdiabatic driving" by Shamreen Iram, Emily Dolson, Joshua Chiel, Julia Pelesko, Nikhil Krishnan, ̈Ozenc Gungor, Benjamin Kuznets-Speck, Sebastian Deffner, Efe Ilker, Jacob G. Scott and Michael Hinczewski
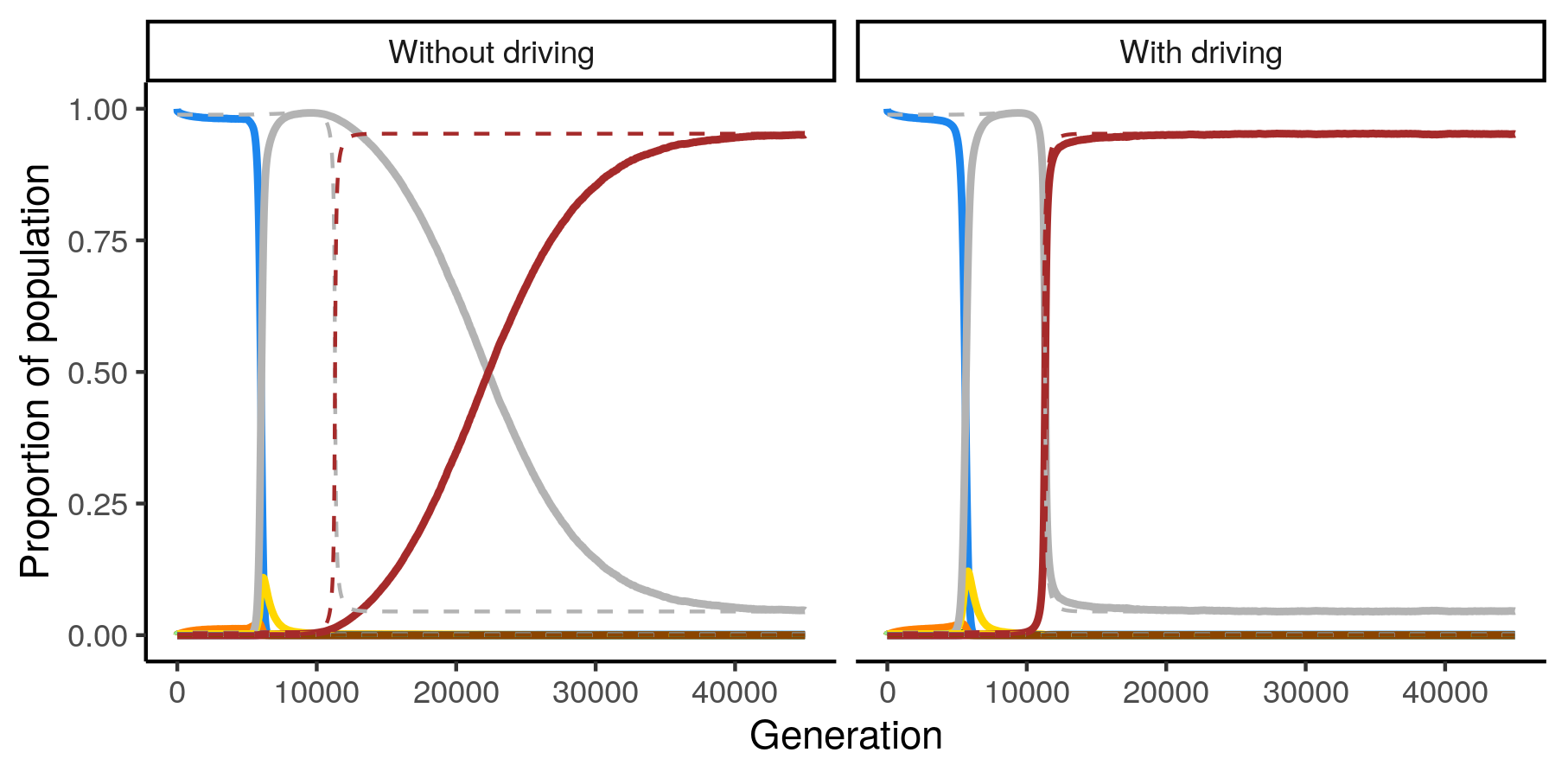 Dashed lines show equilibrium proportion of genotype in population, solid show observed
Dashed lines show equilibrium proportion of genotype in population, solid show observed
- Motivation
- Main takeaway
- Derive your own CD driving prescription (coming soon!)
- Instructions on running the model
- Detailed description of repo contents
Evolution is increasingly recognized as a powerful tool for building and controlling living and lifelike systems. However, the complexity and stochasticity of the evolutionary process have thus far made it more of a blunt instrument than a tool that can be precisely controlled. Here, we present an approach that can begin to give us the level of fine control we need to harness evolution for a wider variety of purposes. A common step in using evolution to achieve a desired end result is to gradually change an environment over time along some continuous axis. Previously, we lacked guidelines for what pattern this environmental change should follow. Here we summarize a recently-developed solution to this problem in the case where we have perfect knowledge of the fitness landscape. Using an approach from physics called counterdiabatic driving, we can mathematically derive a prescription for how to change the environment so that the population arrives at the adaptive end-state as quickly as possible.
Normally, quickly changing the environment a population is evolving in will create lag between the current distribution of genotypes and the distribution of genotypes you would expect to see if you let the population equilibrate to that environment. Using a technique called counterdiabatic driving, we can eliminate this lag!
This animation shows a 16-genotype fitness landscape (composed of all possible combinations of 4 genes that can have one of two alleles). Genotypes connected by lines are mutationally-adjacent. Node size indicates number of individuals in the population with a given genotype. Nodes are depicted with 1) a red circle indicating the population size we would theoretically expect at equilibrium, and 2) a blue circle indicating the observed population size across 1000 runs (there are error bars but they're often too small to see). Where the circles overlap, they look purple. The animation starts with the entire population at genotype 0000, which has none of the four drug-resistance genes*. As the drug concentration** increases, the fitness of genotype 0000 decreases and the population begins to explore the rest of the landscape. Eventually the drug concentration becomes high enough that 1111 becomes fitter than 1110. With counterdiabatic driving, the actual distribution of genotypes in the population shifts almost as fast as the equilibrium value does***. Without counterdiabatic driving it takes a very long time.
* You may notice that the population does not start at the equilibrium value. This is because the fitness peak at 0000 is lower than the one at 1110 and the equilibrium calculation used in this figure did not take into account that the population was all starting at 0000 rather than being spread across the landscape.
** The concentration shown on the figure is the drug concentration in the reference sequence of environments. The actual concentration used in the counterdiabatic driving condition is different.
*** Note that we are using this fitness landscape as an example of a complex real-world fitness landscape to demonstrate that we can use counterdiabatic driving to speed up evolution. We do not endorse using it to speed up the evolution of drug-resistance in the real world unless it's part of a carefully thought-out plan :)
We're still working out the kinks on this but it will be here soon!
First, clone this repository by executing the following command in a terminal:
git clone https://github.com/Peyara/Evolution-Counterdiabatic-Driving.gitThis repository contains a 1-dimensional implementation of the model and an n-dimensional implementation.
The n-dimensional model has a dependency on the Empirical library. To run it, clone that as well:
git clone https://github.com/emilydolson/Empirical.git
cd Empirical
git checkout memic_model # We're using a few features that haven't been merged into master yet
cd ..Then go into the directory for this repository:
cd Evolution-Counterdiabatic-DrivingYou can run the 1D model with the following commands:
make 1d # compile the code
./1_dimension # run the codeYou will be prompted to enter the number of generations to run each simulation for.
You can run the n-dimensional model with:
make nd # compile the code
./n_dimensions # run the code(note that this requires a relatively modern compiler - e.g. g++ 8 or higher)
This model has a few parameters:
- RANDOM_SEED (integer): the seed to initialize the random number generator with (for repeatability)
- GENERATIONS (integer): the number of generations to run for
- N_GENOTYPES (integer): the number of possible genotypes to mutate among (i.e. the number of dimensions to use for this realization of the model)
- K (floating point number): the carrying capacity
- DEATH_RATE (floating point number): The probability of an indiviudal dieing each generation (d in the equations)
- MAX_BIRTH_RATE (floating point number): The maximum possible birth rate (b0 in the equations)
- FITNESSES (string): Each genotype in the model needs a fitness. These fitnesses are expressed relative to a focal genotype (genotype 0), using the s parameter from the equations. Fitnesses can be specified directly on the command line or by providing the name of a file to look them up in. They should be specified as numbers separated by commas (see landscapes/example/relative_fitnesses.dat for an example). The model needs a number of fitnesses equal to the number of genotypes. Example:
-FITNESSES 0,.3,.2. - INIT_POPS (string): Each genotype needs an initial population level. These are specified in the same way as FITNESSES (see landscapes/example/init_pops.dat for an example).
- TRANSITION_PROBS (string): Each pair of genotypes needs probabilities for mutating between those genotypes (the probabilities can be different depending on the direction of the mutation). These probabilities are specified by providing a matrix. The diagonal of the matrix indicates the probabilities of not mutating. As with FITNESSES and INIT_POPS, this can either be provided in line or by referring to a file. The matrix is laid-out such that rows are the genotype being mutated from and columns are the genotype being mutated too. Within a row, numbers should be separated by commas. In a file rows can be separated with new-lines (example in landscapes/example/transition_probs.dat). Because new-lines are hard to type as part of a command, rows should be separated with colons when the matrix is entered on the command line. Example:
-TRANSITION_PROBS .1,.9:.2,.8. The matrix should be square (same number of rows and columns), and have a number of rows and columns equal to the number of genotypes. Numbers within a row should sum to 1 (since offspring need to have one of the available genotypes). - FITNESS_CHANGE_RULE (int): The goal of this model is to explore how we can steer evolution by changing relative fitnesses. There are a variety of different regimes that we might use: 0 (NONE) - relative fitnesses stay constant, 1 (VAR), 2 (VARCD) - a counterdiabatic steering protocol, 3 - Drug with increasing dose, 4 - Drug with fixed dose, and 5 - CD Driving prescription. Options 1 and 2 currently only affect the fitness of the genotype specified by GENOTYPE_TO_DRIVE and ignore initial fitness values.
- GENOTYPE_TO_DRIVE (int): If FITNESS_CHANGE_RULE is 1 or 2, this parameter specifies which genotype's fitness should be changed.
- TIME_STEPS_BEFORE_RAMP_UP (int): If FITNESS_CHANGE_RULE is 3, this parameter specifies how many time steps to wait before increasing drug concentration.
- DRUG_DOSE (double): If FITNESS_CHANGE_RULE is 3 or 4, this parameter specfies the drug dosage to use. For rule 3, this will be the maximum dose that is eventually reached. For rule 4, this will be the single, constant dose that is present for the entire run.
- CD_DRIVING_PRESCRIPTION (string): If FITNESS_CHAGE_RULE is 5, this parameter specifies a file to load the counterdiabatic driving protocol from. The file is expected to have one line for each time step in the model (as specified with the GENERATIONS parameter). Each line should have N_GENOTYPES values representing relative fitnesses of each genotype at each time step. Values should be separated with commas.
- IC50S (string): Needed for FITNESS_CHANGE_RULE 3 and 4. The IC50 values for each genotype, which are used to determine each genotype's fitness at a given drug concentration, based on the equation presented by Ogbunugafor et. al. Specified in the same format as FITNESSES (either a list of comma-separated values or the name of a file containing comma-separated values).
- G_DRUGLESSES (string): Needed for FITNESS_CHANGE_RULE 3 and 4. The growth rates for each genotype in the absence of drug, which are used to determine each genotype's fitness at a given drug concentration, based on the equation presented by Ogbunugafor et. al. Specified in the same format as FITNESSES (either a list of comma-separated values or the name of a file containing comma-separated values).
- CS (string): Needed for FITNESS_CHANGE_RULE 3 and 4. The equation we use to calculate fitnesses at various drug concentrations has a fitting parameter, C. Usually its value is the same for all genotypes, but this model allows different values to be specified per-genotype if desired. Use this parameter to specify its value, using the same format as FITNESSES (either a list of comma-separated values or the name of a file containing comma-separated values).
The values of these can be set with command line flags by placeing a dash before the name of the parameter you would like to modify and following it with the desired parameter value:
# For example, the following runs the model with a DEATH_RATE of .1 and GENERATIONS set to 100
./n_dimensions -DEATH_RATE .1 -GENERATIONS 100Alternatively, they can be set by modifying the values in the configuration file, NDim.cfg. The model will use any file called NDim.cfg in the current directory with it as a configuration file. The configuration file used for this paper is stored in the config directory, but will be copied to the current directory when you run make nd.
config/NDim.cfg contains the base configuration, without counterdiabatic driving. Parameter modifications used to run other conditions were specified as command-line arguments. For the exact values used to generate data in the paper, see the SLURM submission scripts in the config directory.
All code for both models lives in the source directory. The 1-dimensional model is in ABMtoFP_Evol.c. The majority of the code for the n-dimensional model is in n_dimensions.h and the remainder is in n_dimensions.cc.
The three driving prescriptions used in the paper are in the driving_prescriptions directory. The numbers after maxc in the filenames indicate the maximum allowed drug concentration in each. These prescriptions where derived as explained in the paper. These prescriptions are designed specifically for the pyrimethamine resistance fitness landscape with the drug increase profile described by FITNESS_CHANGE_RULE 3, a mutation rate of .001, and a carrying capacity of 5,000,000.
The landscapes directory contains configuration files to run the model on various fitness landscapes. The only fully-specified fitness landscape currently here is the pyrimethamine resistance fitness landscape from Ogbunugafor et. al, in the landscapes/malaria_landscapes subdirectory. It contains initial population sizes, log10(IC50) values, initial fitnesses (expressed as s, i.e. relative fitness), fitnesses in the absence of drug (expressed as growth rate - not relative fitness), and values of the constant c.
There is also a landscapes/ecoli_landscapes directory. These contain files with the fitnesses of 16 E. coli genotypes when subjected to different antibiotics (from this repository). We can't currently use these in the model because we need IC50 and drugless growth rate value, but they are a promising avenue for future investigation.
The landscapes/mutation_matrices directory contains mutation matrices to supply to the TRANSITION_PROBS parameter. Each matrix assumes all mutations to adjacent (i.e. Hamming distance 1) genotypes are equally likely and no other mutations are possible. The files in this directory are mutation matrices for 16-genotype fitness landscapes with the mutation rate specified in the file name (e.g. mut_matrix_.01.dat has a mutation rate of .01). The mutation rate refers to the total probability of an offspring having a different genotype than its parent.
The landscapes/example directory contains example configuration files.
Lastly, the landscapes/scripts directory contains a few useful scripts for working with these landscapes and creating new ones:
make_trans_prob.pyis a script that takes the number of genotypes in the landscape and the mutation rate as command-line arguments, and outputs the mutation matrix resulting from those parameters.convert_fitness_to_s.pyis a script for converting from absolute fitness (e.g. growth rate) to the relative fitness values (s) used in this model. It takes the name of a file containing a single line of comma-separated numbers as input and alters the values in that file. WARNING: overwrites the data in the file you give it - make a backup!plot_curves.pyis a script that reproduces the pyrimethamine dose-fitness curves from Ogbunugafor et. al, as a check that we're doing the math right.