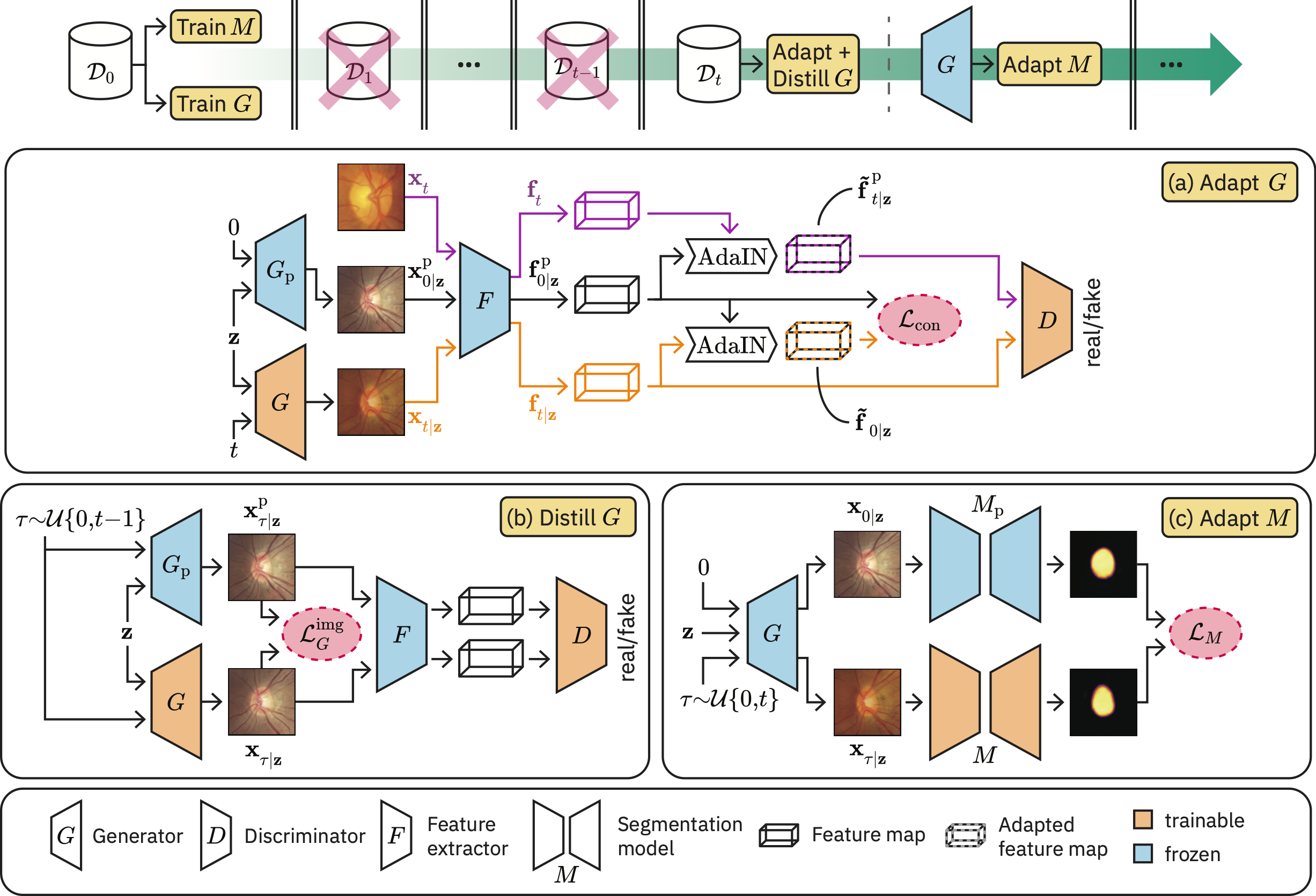
Generative Appearance Replay is a novel segmentation method for strictly continual domain adaptation wherein no access to the source domain or other previously seen domains is required. This repository contains the PyTorch code of our corresponding paper Generative appearance replay for continual unsupervised domain adaptation.
Create a conda environment and install the required packages from the provided environment.yml:
git clone https://github.com/histocartography/generative-appearance-replay
cd generative-appearance-replay
conda env create -f environment.yml
conda activate garda
Install pytorch and torchvision:
conda install pytorch==1.10.1 torchvision==0.11.2 -c pytorch
We evaluated our method for three different segmentation tasks:
-
Optic disc segmentation (ODS):
- Datasets:
- Preprocessing adopted from this repository
- train/val/test splits and lists of all images can be found under
doc/ods_splits/
-
Cardiac segmentation (CS):
- Images of 3 different vendors from the M&Ms dataset
- Datasets:
- Source domain: vendor B
- Target domain 1: vendor A
- Target domain 2: vendor D
- Preprocessing adapted from this repository. We added bias field correction, resized the images to 256x256 instead of 224x224, used "constant" instead of "edge" padding, and saved the ground truth masks as 1-channel .npy matrices with values 0 (background), 1 (left ventricle myocardium), 2 (left ventricle), and 3 (right ventricle)
- train/val/test splits and lists of all images can be found under
doc/cs_splits/
-
Prostate segmentation (PS):
- Images of 6 different sites from the Multi-site Prostate Segmentation dataset
- Datasets:
- Source domain = RUNMC (renamed to Site_A)
- Target domain 1 = BMC (renamed to Site_B)
- Target domain 2 = I2CVB (renamed to Site_C)
- Target domain 3 = UCL (renamed to Site_D)
- Target domain 4 = BIDMC (renamed to Site_E)
- Target domain 5 = HK (renamed to Site_F)
- Preprocessing can be found in
bin/preproc_ps.py(adapted from this repository) - train/val/test splits and lists of all images can be found under
doc/ps_splits/
Below we provide example instructions on how to run preprocessing, as well as how to adapt a segmentation model and a GAN for prostate segmentation over a sequence of domains, given the sample config files under source/config/ps/.
Preprocess the data downloaded from https://liuquande.github.io/SAML/ :
python bin/preproc_ps.py --in_path /path/to/data/raw/ --out_path /path/to/data/preprocessed/
This will create the following directory structure:
/path/to/data/preprocessed/
├── Site_A/
└── all_slides.csv
└── split.csv
└── images/
└── Case00_Site_A_slice00.png
└── Case00_Site_A_slice01.png
:
└── masks/
└── Case00_Site_A_slice00.npy
└── Case00_Site_A_slice01.npy
:
├── Site_B/
:
├── Site_F/
-
Train the segmentation model on the source domain:
python bin/run.py --data_path /path/to/data --config_path source/config/ps/src_seg_train.json --task ps -
Train the GAN on the source domain:
python bin/run.py --data_path /path/to/data --config_path source/config/ps/src_gen_train.json --task ps -
Adapt the GAN to a target domain:
python bin/run.py --data_path /path/to/data --config_path source/config/ps/tgt1_gen_train.json --task ps -
Adapt the segmentation model to a target domain:
python bin/run.py --data_path /path/to/data --config_path source/config/ps/tgt1_seg_train.json --task ps
Steps 3 and 4 are then repeated accordingly for every additional target domain.
To test the model, set the config field "task" to "test_seg" and provide the path of the model to be tested in the field "seg_model_path", as shown in source/config/ps/tgt5_seg_test.json.
If you use this code, please make sure to cite our work:
@article{chen2023garda,
title = {Generative appearance replay for continual unsupervised domain adaptation},
journal = {Medical Image Analysis},
volume = {89},
pages = {102924},
year = {2023},
author = {Boqi Chen and Kevin Thandiackal and Pushpak Pati and Orcun Goksel},
}