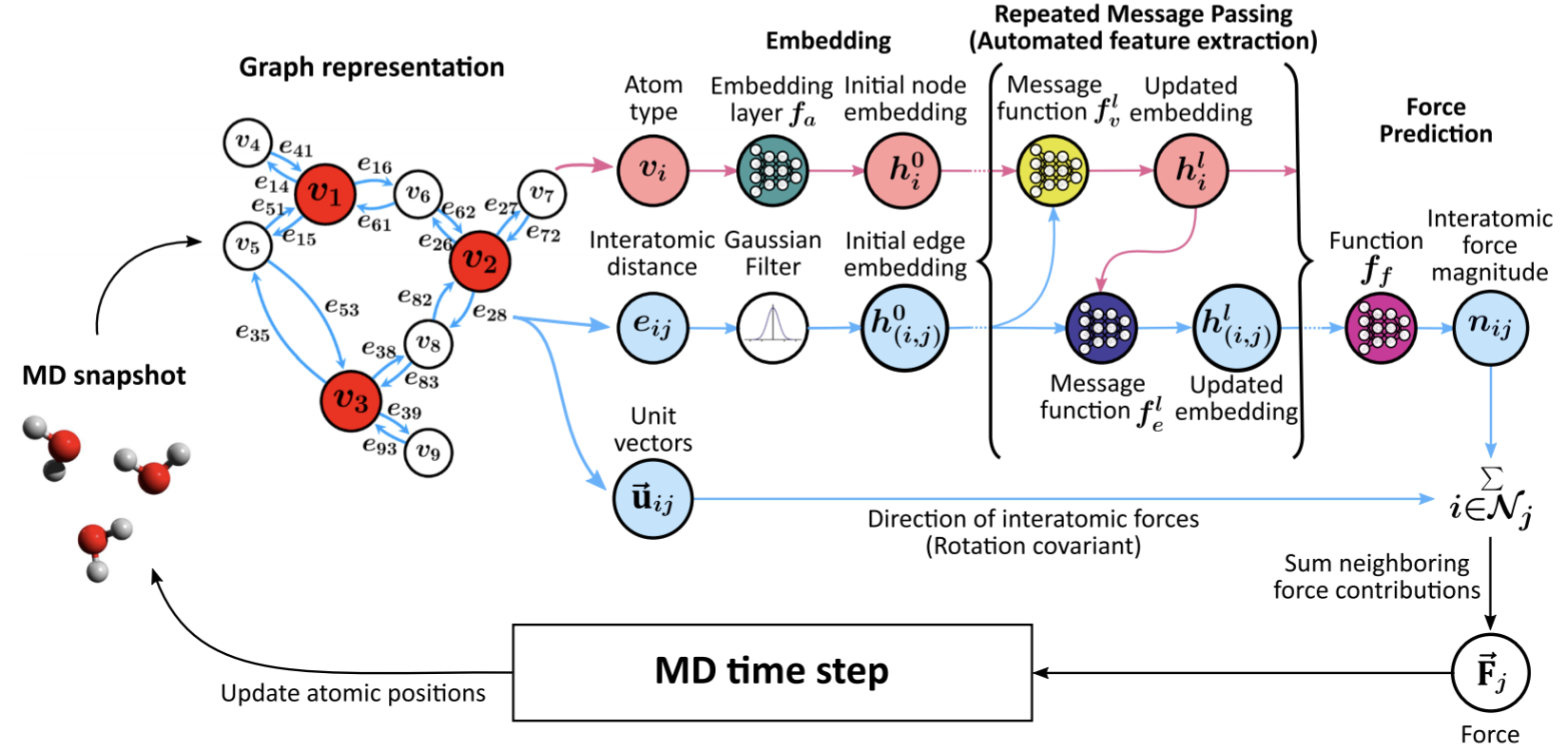
Atomistic Force Fields based on GNNFF
GNNFF [1] is a graph neural network framework to directly predict atomic forces from automatically extracted features of the local atomic environment that are translationally-invariant, but rotationally-covariant to the coordinate of the atoms. This package is an atomistic force fields that constructed based on GNNFF.
Requirements:
- python 3
- ASE
- numpy
- PyTorch (>=1.7)
- schnetpack
Note: We recommend using a GPU for training the neural network.
Install this project by git.
git clone https://github.com/ken2403/gnnff.git
cd gnnffpip install -r requirements.txtpip install .You're ready to go!
The example scripts provided by gnnff are inserted into your PATH during installation. These scripts are in gnnff/src/scripts.
Prepare the dataset in ase database format.
Or prepare a atoms .xyz file and convert it to ase database using the script provided by schnetpack.
spk_parse.py /path_to_xyz /save_path_to_dbPrepare the .json file that defines the hyperparameters for training.
The example file can be found in gnnff/src/scripts/argument
If the argument .json file is ready, you can train the model like this way.
gnnff_run from_json /path_to_train.jsonLearned model are saved as best_model in model direcctory.
The same script can also be used to evaluate the learned model.
Prepare a .json file for evaluation.
gnnff_run from_json /path_to_eval.jsonWrite a result file evaluation.txt into the model directory.
You can also input one POSCAR structure file into the learned model to calculate total energy and interatomic forces.
This is useful for phonon calculations with phonopy.
gnnff_run from_poscar /path_to_POSCAR /path_to_learned_model --cutoff cutoff_radious [--cuda]If GPU is availabel, set --cuda.
The calculation results are written to gnnff_run.xml file and saved in the same directory as the POSCAR.
-
[1] C. W. Park, M. Kornbluth, J. Vandermause, C. Wolverton, B. Kozinsky, J. P. Mailoa, Accurate and scalable graph neural network force field and molecular dynamics with direct force architecture. npj Computational Materials. 7, 1–9 (2021). link
-
[2] T. Xie, J. C. Grossman, Crystal Graph Convolutional Neural Networks for an Accurate and Interpretable Prediction of Material Properties. Phys. Rev. Lett. 120, 145301 (2018). link
-
[3] implementaion of Crystal Graph Convolutional Neural Networks