- Objective
- How?
- Requirements
- Execution examples
- Example output
- Extend a calculation with more genomes
- A note on alignment calculation (DIAMOND vs. BLASTP) and benchmarking
- Parameter adjustments (danger zone)
- All-vs-All and One-vs-All
- Cite
- Update backlog
Sequence technology advancements have led to an exponential increase in bacterial genomes, necessitating robust taxonomic classification methods. The Percentage Of Conserved Proteins (POCP), proposed initially by Qin, Xie et al. 2014, is a valuable metric for assessing prokaryote genus boundaries. A prokaryotic genus can be defined as a group of species with all pairwise POCP values higher than 50%. Here, I introduce a computational pipeline for automated POCP calculation, aiming to enhance reproducibility and ease of use in taxonomic studies (see publication in OUP Bioinformatics).
As input use one amino acid sequence FASTA file per genome such as provided by Prokka or Bakta or genome FASTA files which will be then annotated via Prokka automatically. The pipeline will then calculate all-vs-all pairwise alignments between all protein sequences using the blastp mode of DIAMOND and use this information for POCP calculation following the original formula of Qin, Xie et al. 2014. One-vs-all comparisons are also possible, see below.
You only need nextflow to install and run the pipeline. nextflow will take care of all dependencies and install them if necessary. For installing the dependencies (such as Prokka and DIAMOND), you can choose between conda, mamba, docker or singularity. I recommend using docker. Then install and run the pipeline:
Use the -profile parameter to switch between different dependency managers, e.g. -profile conda.
# get the pipeline code
nextflow pull hoelzer/pocp
# check availble release versions and development branches, recommend to use latest release
nextflow info hoelzer/pocp
# get the help page and define a release version. ATTENTION: use latest version.
nextflow run hoelzer/pocp -r 2.3.0 --help
# example with genome files as input, all-vs-all comparison, performing a local execution and using Docker
nextflow run hoelzer/pocp -r 2.3.0 --genomes $HOME'/.nextflow/assets/hoelzer/pocp/example/*.fasta' -profile local,docker
# example with protein FASTA files as input (e.g. from Prokka pre-calculated), all-vs-all comparison, performing a SLURM execution and using conda
nextflow run hoelzer/pocp -r 2.3.0 --proteins $HOME'/.nextflow/assets/hoelzer/pocp/example/*.faa' -profile slurm,conda
# example with genome files as input, additional genome file to activate one-vs-all comparison, performing a local execution and using Docker
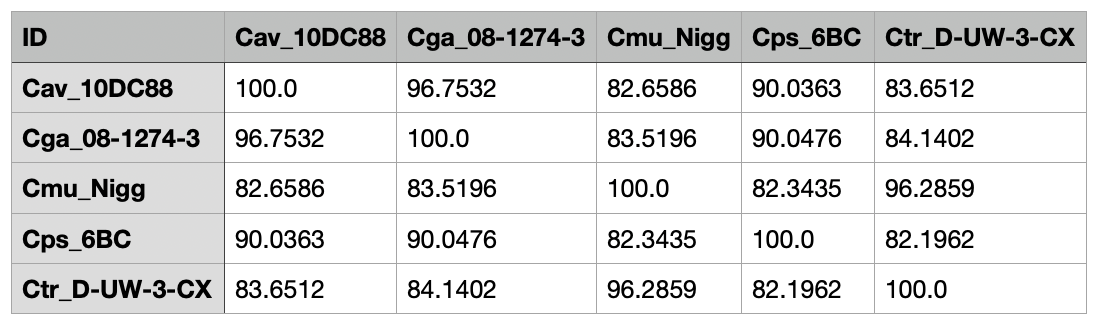
nextflow run hoelzer/pocp -r 2.3.0 --genomes $HOME'/.nextflow/assets/hoelzer/pocp/example/*.fasta' --genome $HOME/.nextflow/assets/hoelzer/pocp/example/Cav_10DC88.fasta -profile local,dockerThe final output (pocp-matrix.tsv) should look like this (here, the resulting TSV was imported into Numbers on MacOS):
nextflow allows you to resume a calculation without recalculating previous steps. This is possible, because nextflow stores intermediate and results files in a work directory. If you want to resume a calculation later, dont delete the work directory and use the same nextflow run command. For example:
# first iteration
nextflow run hoelzer/pocp -r 2.3.0 --genomes 'input/*.fasta' -profile local,docker
# add more genomes to the input folder:
cp /some/other/genomes/*.fasta input/
# resume the calculation via adding "-resume".
nextflow run hoelzer/pocp -r 2.3.0 --genomes 'input/*.fasta' -profile local,docker -resumeThe pipeline identifies orthologous proteins between species using DIAMOND in blastp mode. Please note that the original POCP publication used BLASTP for calculating the alignments. However, DIAMOND is not only faster, which is an advantage when calculating POCP values for larger input data sets, but also achieves the sensitivity of BLASTP (Buchfink (2021)), especially when using the --ultra-sensitive mode, which is activated by default in the pipeline. Another study comparing different alignment programs found that DIAMOND offered the best compromise between speed, sensitivity and quality when a sensitivity option other than the default setting was selected (Hernández-Salmerón and Moreno-Hagelsieb (2020)). Therefore I decided to use DIAMOND as a more modern solution for the alignment calculation in POCP-nf. Thx @michoug for the idea and the Pull Request.
Using five bacteria data sets spanning 15 to 167 input genomes, I calculated that the average difference in percentage of POCP scores is ~0.16 % between using BLASTP or DIAMOND in --ultra-sensitive mode. The runtime (without Prokka annotation, --proteins input) for 44 Enterococcus genomes (comprising 1,892 pairwise comparisons) is halved from 10h 12m (BLASTP) to 5h 30m (DIAMOND) on a laptop with 8 cores.
If you are interested in more details, check out this little benchmark.
Attention: Although you can customize core parameters in the nextflow pipeline, I recommend sticking to the original parameters defined by Qin, Xie et al. 2014 and otherwise clearly indicating any changed parameter options along with the version of POCP-nf used when sharing POCP results! The default parameters defined in the configuration of the pipeline are:
--evalue 1e-5
--seqidentity 0.4
--alnlength 0.5A warning will be shown by the pipeline if you change these parameters.
Please also note that per default an "all-vs-all" comparison is performed based on the provided FASTA files. However, you can also switch to an "one-vs-all" comparison by additionally providing a single genome FASTA via --genome next to the --genomes input or a single protein multi-FASTA via --protein next to the --proteins input. In both cases, only "one-vs-all" comparisons will be performed. It is also possible to combine --genomes with a target --protein FASTA for "one-vs-all" and vice versa.
If you use the POCP Nextflow pipeline, please cite the original POCP study that introduced the metric, the publications of the great tools used in the pipeline, and the POCP-nf pipeline:
Update 2023/12: One-vs-All comparisons are now possible in genome and protein input mode. Check --help message.
Update 2023/10: Now using Diamond instead of Blastp for protein alignments. Thx @michoug for the Pull Request.
Update 2023/05: Re-implementation as a Nextflow pipeline. Please feel free to report any issues!