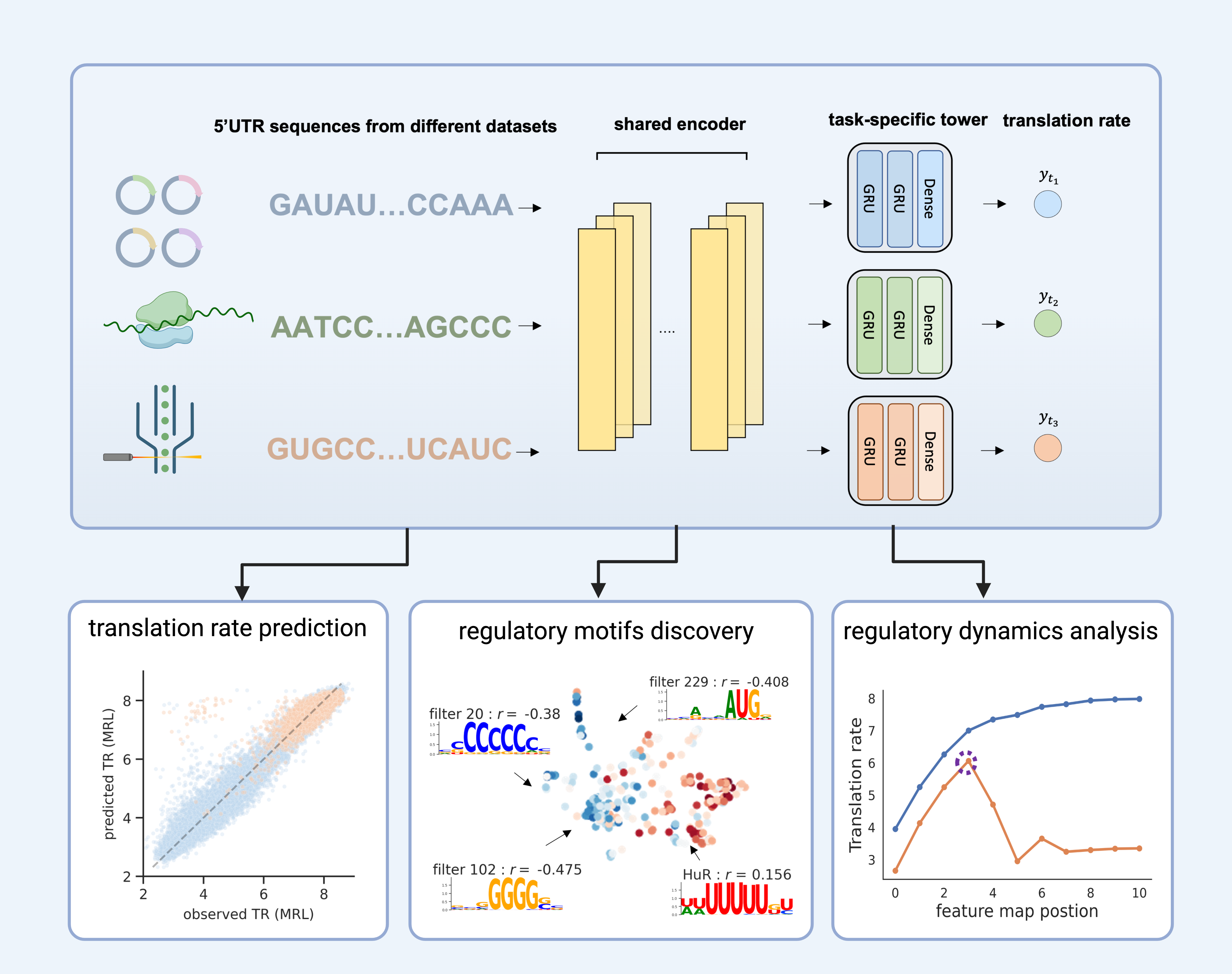
MTtrans is a multi-task learning framework for integrating translation rate measurement from differet sequencing platforms and the discovery. The shared prepresentation captures common patterns acorss techiniques that are used to discover the robust cis-regulatory elements.
MTtrans is implemented with pytorch.
We use conda to manage the python packages used in this study, which
is required for setting up the environment. If you don't have conda installed in your machine, please refer to conda guide.
# clone to local and enter the main directory
git clone https://github.com/holab-hku/MTtrans
cd MTtrans
# make sure you are stay under the main folder
bash script/generate_config_js.sh
# create a new environment and install the prerequisites
conda env create -f util/environment.yml
conda activate MTtransAfter running the shell script script/generate_config_js.sh, you are supposed to see file machine_configure.json
under the main directory. This file locates the codes and the data that will be downloaded later.
Please don't change the "script_dir".
The "data_dir" and "pth_dir" record where the datasets and the models will be saved. The default setting is data/ and checkpoint/ under the this main folder.
There are 7 datasets used in total, including 3 massively parallel report assay (MPA), 3 ribosome profilings (RP) and 1 MPA dataset performed in the yeast system. You can download them by
# make sure you are stay under the main folder
bash script/download_training_data.sh Then data preprocessing by
bash script/data_prepocessing.shThe processed files will all saved to the "data_dir" defined in the machine_configure.json.
Under the terminal, we can call the python script iter_train.py to impement MTtrans. All the hyper-parameters and other experimental details like task combination are storaged in the configuration file in the format of ini. Let's have a quick tesing run by
# an one-epoch testing experiment
python script/iter_train.py --config_file log/Backbone/RL_hard_share/3R/testing.iniIf the python script works, you will see the following text popping both in the shell and the
- ini file updated
- Snapshot saved to <pth_dir>/RL_hard_share_MTL/3R/testing-model_best_cv1.pthThe testing model will automatically save to the "pth_dir", which is checkpoint/ by default.
To explore the trained model, we can load in the saved checkpoint to resume the model for later analysis.
Here we use the testing checkpoint file (end with .pth) we just trained as the example.
The checkpoints of all kinds of our MTtrans variant is available at https://figshare.com/articles/dataset/MTtrans_Model_weight/24504154.
# firstly make sure you are in the correct environment
conda activate MTtrans
# then get into your favourite python IDE (i.e jupyter / vscode)
# here I enter the built-in one for demonstration
python>>> import os
>>> import torch
>>> from torchinfo import summary
# paste the checkpoint path here
>>> save_path = "<pth_dir>/RL_hard_share_MTL/3R/testing-model_best_cv1.pth"
>>> assert os.path.exists(save_path) , "please repalce <pth_dir> with the correct path set in your machine"
# resume in the 'cpu'
>>> model = torch.load(save_path, 'cpu')['state_dict']
# an overview of the model
>>> summery(model)
=====================================================================
Layer (type:depth-idx) Param #
======================================================================
RL_hard_share --
├─Conv1d_block: 1-1 --
│ └─ModuleList: 2-1 --
│ │ └─Sequential: 3-1 1,920
│ │ └─Sequential: 3-2 99,072
│ │ └─Sequential: 3-3 394,752
│ │ └─Sequential: 3-4 787,968
├─ModuleDict: 1-2 --
│ └─ModuleList: 2-2 --
│ │ └─GRU: 3-5 181,440
│ │ └─Linear: 3-6 81
│ └─ModuleList: 2-3 --
│ │ └─GRU: 3-7 181,440
│ │ └─Linear: 3-8 81
│ └─ModuleList: 2-4 --
│ │ └─GRU: 3-9 181,440
│ │ └─Linear: 3-10 81
│ └─ModuleList: 2-5 --
│ │ └─GRU: 3-11 181,440
│ │ └─Linear: 3-12 81
│ └─ModuleList: 2-6 --
│ │ └─GRU: 3-13 181,440
│ │ └─Linear: 3-14 81
├─Linear: 1-3 81
├─MSELoss: 1-4 --
======================================================================
Total params: 2,191,398
Trainable params: 2,191,398
Non-trainable params: 0
======================================================================
>>>exit()We sincerely thanks all the listed authors for making their data publicly available.
Cuperus, J.T., Groves, B., Kuchina, A., Rosenberg, A.B., Jojic, N., Fields, S., Seelig, G.: Deep learning of the regulatory grammar of yeast 5 untranslated regions from 500,000 random sequences. Genome Research 27(12), 2015{2024 (2017) link
Sample, P.J., Wang, B., Reid, D.W., Presnyak, V., McFadyen, I.J., Morris, D.R., Seelig, G.: Human 5 utr design and variant effect prediction from a massively parallel translation assay. Nature Biotechnology 37(7), 803{809 (2019) link
Andreev, D.E., O’Connor, P.B., Fahey, C., Kenny, E.M., Terenin, I.M., Dmitriev, S.E., Cormican, P., Morris, D.W., Shatsky, I.N., Baranov, P.V.: Translation of 5 leaders is pervasive in genes resistant to eif2 repression. Elife 4, 03971 (2015) link
Hsieh, A.C., Liu, Y., Edlind, M.P., Ingolia, N.T., Janes, M.R., Sher, A., Shi, E.Y., Stumpf, C.R., Christensen, C., Bonham, M.J., et al.: The translational landscape of mtor signalling steers cancer initiation and metastasis. Nature 485(7396), 55{61 (2012) link
Wein, N., Vulin, A., Falzarano, M.S., Szigyarto, C.A.-K., Maiti, B., Findlay, A., Heller, K.N., Uhl´en, M., Bakthavachalu, B., Messina, S., et al.: Translation from a dmd exon 5 ires results in a functional dystrophin isoform that attenuates dystrophinopathy in humans and mice. Nature Medicine 20(9), 992{1000 (2014) link
Cao, J., Novoa, E.M., Zhang, Z., Chen, W.C., Liu, D., Choi, G.C., Wong, A.S., Wehrspaun, C., Kellis, M., Lu, T.K.: High-throughput 5 utr engineering for enhanced protein production in non-viral gene therapies. Nature Communications 12(1), 1{10 (2021) link