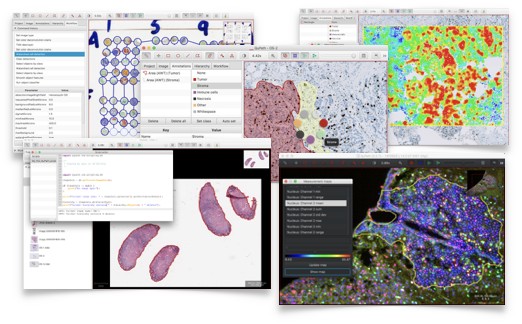
QuPath is open source software for whole slide image analysis and digital pathology.
QuPath has been developed as a research tool at Queen's University Belfast. It offers a wide range of functionality, including:
- extensive tools for annotation and visualization
- workflows for both IHC and H&E analysis
- novel algorithms for common tasks, e.g. cell segmentation, Tissue microarray dearraying
- interactive machine learning, e.g. for cell and texture classification
- an object-based hierarchical data model, with scripting support
- extensibility, to add new features or support for different image sources
- easy integration with other tools, e.g. MATLAB and ImageJ
All in all, QuPath aims to provide researchers with a new set of tools to help with bioimage analysis in a way that is both user- and developer-friendly.
QuPath is free and open source, using GPLv3.
To download a version of QuPath to install, go to the Latest Releases page.
For documentation and more information, see the QuPath Wiki
Copyright 2014-2016 The Queen's University of Belfast, Northern Ireland
- Pete Bankhead
- Jose Fernandez
- Prof Peter Hamilton
- Prof Manuel Salto-Tellez
The QuPath software has been developed as part of projects that have received funding from:
- Invest Northern Ireland (RDO0712612)
- Cancer Research UK Accelerator (C11512/A20256)
To see a list of third-party open source libraries used within QuPath (generated automatically using Maven), see THIRD-PARTY.txt.
Full licenses and copyright notices for third-party dependencies are also included for each relevant submodule inside src/main/resources/licenses, and accessible from within within QUPath distributions under Help → License.