⚡️⚡️⚡️ Winner of CIKM 2022 Best Paper
D-HYPR: Harnessing Neighborhood Modeling and Asymmetry Preservation for Digraph Representation Learning
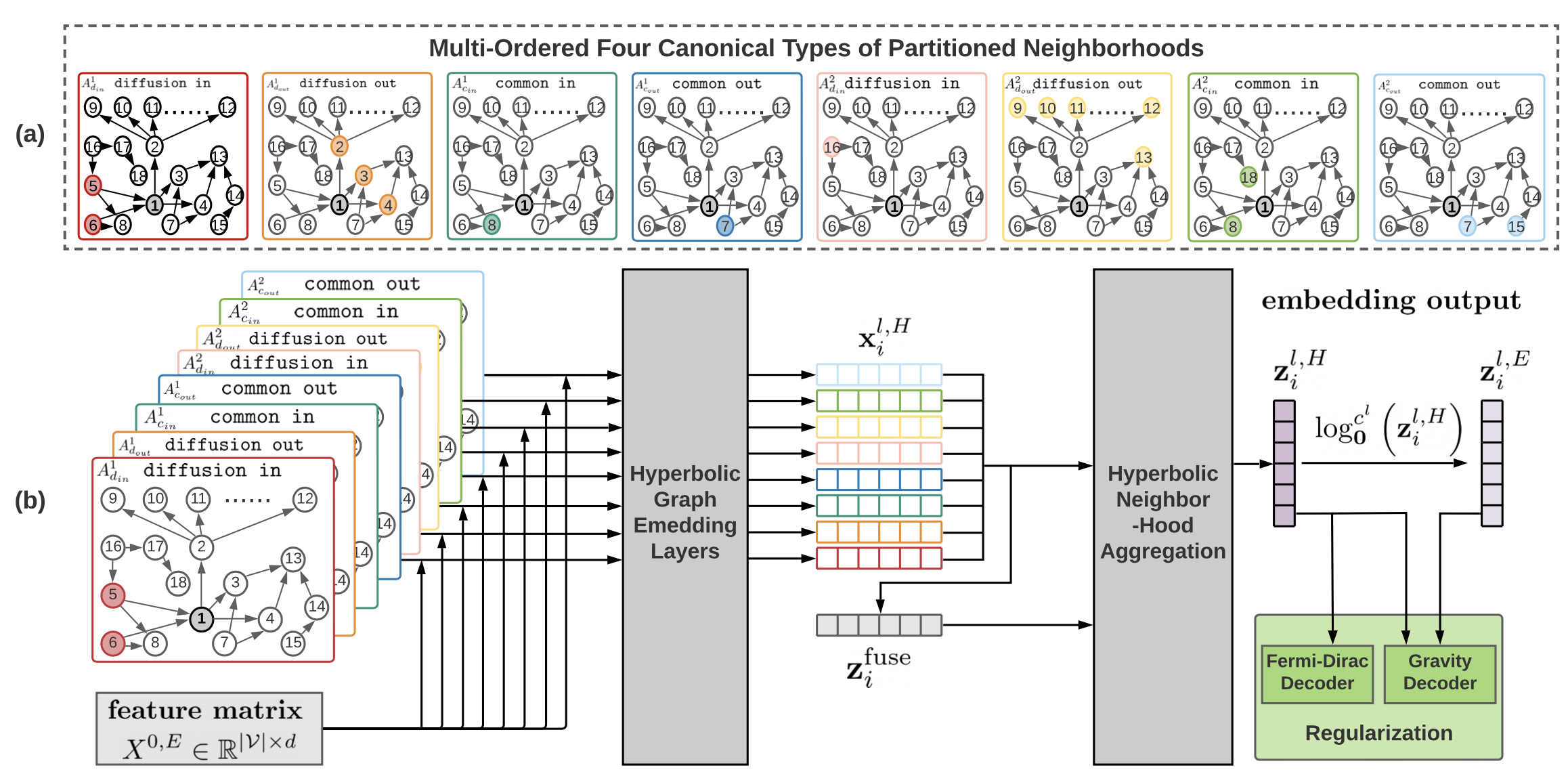
This is an official implementation of the CIKM 2022 paper D-HYPR: Harnessing Neighborhood Modeling and Asymmetry Preservation for Digraph Representation Learning. In this repository, we provide the PyTorch code for training and evaluation as described in the paper. The proposed model Digraph Hyperbolic Networks (D-HYPR) considers the unique node neighborhoods in digraphs with multi-scale neighborhood collaboration in hyperbolic space. D-HYPR respects asymmetric relationships of node-pairs, which is guided by sociopsychology-inspired regularizers.
D-HYPR outperforms SOTA on multiple downstream tasks including node classification, link presence prediction, and link property prediction. Particularly, D-HYPR generates meaningful embeddings in very low dimensionalities.
If you find our repo useful in your research, please use the following BibTeX entry for citation.
@article{zhou2022dhypr,
title={D-HYPR: Harnessing Neighborhood Modeling and Asymmetry Preservation for Digraph Representation Learning},
author={Zhou, Honglu and Chegu, Advith and Sohn, Samuel S and Fu, Zuohui and De Melo, Gerard and Kapadia, Mubbasir},
journal={Proceedings of the 31st ACM International Conference on Information and Knowledge Management (CIKM 2022)},
year={2022}
}conda env create -f environment.ymlPlease download the data folder using this link.
data/
├── air
├── blog
├── citeseer
├── cora
├── cora_ml
├── dblp
├── survey
├── wiki
The folds of each dataset are provided. Each fold is a unique train-test split of the dataset.
The scripts to run D-HYPR on the 8 datasets for node classification, link presence prediction, and link sign prediction are available in dhypr/code/D-HYPR.
To get started with a one-time experiment:
dhypr/code/dhypr$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_lp_air.sh To obtain the results on all folds with repeated runs (100 runs in total using different seeds):
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python lp_air.pyTo get started with a one-time experiment:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_lp_cora.sh To obtain the results on all folds with multiple runs (100 runs in total using different seeds):
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python lp_cora.pyTo get started with a one-time experiment:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_lp_blog.sh To obtain the results on all folds with multiple runs (100 runs in total using different seeds):
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python lp_blog.pyTo get started with a one-time experiment:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_lp_survey.sh To obtain the results on all folds with multiple runs (100 runs in total using different seeds):
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python lp_survey.pyTo get started with a one-time experiment:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_lp_dblp.sh To obtain the results on all folds with multiple runs (100 runs in total using different seeds):
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python lp_dblp.pyTo get started with a one-time experiment:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_nc_cora_ml.sh To obtain the results on all folds:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python nc_cora_ml.pyTo get started with a one-time experiment:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_nc_citeseer.sh To obtain the results on all folds:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python nc_citeseer.pyTo get started with a one-time experiment:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_nc_wiki.sh To obtain the results on all folds:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python nc_wiki.pyTo get started with a one-time experiment:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_vnc_cora_ml.sh To obtain the results on all folds:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python vnc_cora_ml.pyTo get started with a one-time experiment:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_vnc_citeseer.sh To obtain the results on all folds:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python vnc_citeseer.pyTo get started with a one-time experiment:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_vnc_wiki.sh To obtain the results on all folds:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python vnc_wiki.pyTo get started with a one-time experiment:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ ./onefold_sp_wiki.sh To obtain the results on all folds:
dhypr/code/D-HYPR$ conda activate dhypr
dhypr/code/D-HYPR$ python sp_wiki.pyPlease use dhypr/code/D-HYPR/collect_result_lp.ipynb to gather the metric result values from the multiple link prediction experiments, and then use dhypr/helper/get_lp_result_avg_std.ipynb to get the average metric result values.
Similarly, dhypr/code/D-HYPR/collect_result_nc.ipynb for node classification, dhypr/code/D-HYPR/collect_result_vnc.ipynb for semi-supervised node classification, and dhypr/code/D-HYPR/collect_result_sp.ipynb for link sign prediction.
The following is an example to demonstrate the entire trainining and evaluation pipeline:
dhypr$ cd code/D-HYPR
dhypr/code/D-HYPR$ conda activate dhyprRun D-HYPR on Cora Link Prediction on just fold 0:
dhypr/code/D-HYPR$ ./onefold_lp_cora.sh Run D-HYPR on Cora Link Prediction (100 times):
dhypr/code/D-HYPR$ python lp_cora.py- Run
collect_result_lp.ipynb cd ../../helper- Run
get_lp_result_avg_std.ipynb
The scripts to run the baselines (MLP, HNN, GCN, GAT, HGCN) on the 8 datasets for node classification, link presence prediction, and link sign prediction are available in dhypr/code/baselines.
The baseline code is organized in a similar fashion as D-HYPR, therefore please refer to the above D-HYPR related train/eval instructions to run the baseline code.
For other baselines such as NERD, ATP and APP, please contact Advith Chegu: ac1771@scarletmail.rutgers.edu
The code of D-HYPR and the code of baselines are heavily based on the following publically available implementations (we sincerely appreciate the authors of these repositories):
- https://github.com/HazyResearch/hgcn
- https://github.com/ydzhang-stormstout/HAT
- https://github.com/flyingtango/DiGCN
- https://github.com/deezer/gravity_graph_autoencoders
- https://github.com/matthew-hirn/magnet
- https://github.com/zhenv5/atp
- https://github.com/AnryYang/APP
- https://git.l3s.uni-hannover.de/khosla/nerd
If you are interested to obtain the four types of k-order proximity matrix described in the paper, please refer to the scripts in the dhypr/preprocessing folder. Please note that the maximal order K (K_max) is currently set to 2. The four types of k-order proximity matrix of the 8 datasets were already extracted and provided in the data foler.