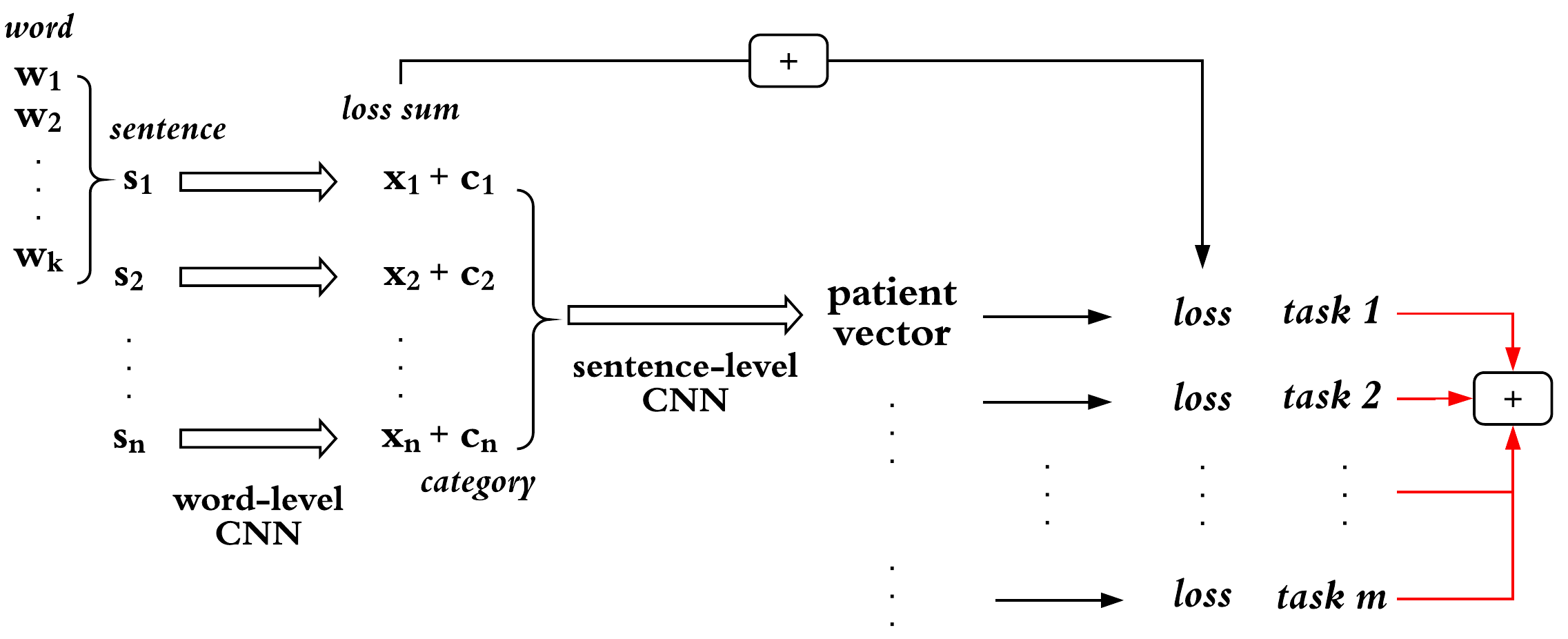
We developed a deep learning-based model for multi-task learning task from ICU patient clinical notes to predict patient mortality and length of stay.
This repository contains the code for this project, also contains the slide and the paper for more details.
Clinical notes are fetched from a public ICU patient database, also known as MIMIC-III. For more information, like how to get access to the data, please refer to MIMIC Critical Care Database.
We don't provide the original data or trained embeddings from MIMIC-III in this repo as it violates the Data Usage Agreement (DUA) of MIMIC-III data. We only provides the instruction for where to download the data: the clinical notes are in the table NOTEEVENTS.csv. The records about admission visits are documented in ADMISSIONS.csv, and patient information is in PATIENTS.csv.
The proposed model in this study, a double-level Convlutional Neural Network (CNN), is illustrated as the following figure:
We propose a deep learning-based multi-task learning (MTL) architecture focusing on patient mortality predictions from clinical notes. The MTL framework enables the model to learn a patient representation that generalizes to a variety of clinical prediction tasks. Moreover, we demonstrate how MTL enables small but consistent gains on a single classification task (e.g., in-hospital mortality prediction) simply by incorporating related tasks (e.g., 30-day and 1-year mortality prediction) into the MTL framework. To accomplish this, we utilize a multi-level Convolutional Neural Network (CNN) associated with a MTL loss component. The model is evaluated with 3, 5, and 20 tasks and is consistently able to produce a higher-performing model than a single-task learning (STL) classifier. We further discuss the effect of the multi-task model on other clinical outcomes of interest, including being able to produce high-quality representations that can be utilized to great effect by simpler models. Overall, this study demonstrates the efficiency and generalizability of MTL across tasks that STL fails to leverage.
- Environment Setting Up
- Directory Structure
- Demo
- Parameters
- Results
- Contact Information
The following instructions are based on Ubuntu 14.04 and Python 3.6, on GPU support.
There are three options to install required environments for running the project:
-
Manual installation (Not recommended but easier to get a basic overview of required packages):
a) Install CUDA toolkit and corresponding cuDNN(https://developer.nvidia.com/cudnn) for GPU support
b) Install Tensorflow. It is a extremely tough experience to install GPU support Tensorflow on Ubuntu but always remember to refer to the official guidence.
pip3 install tensorflowandpip3 install tensorflow-gpuc) Additional packages are needed: logging (tracking events that happen, similar as print) and tqdm (progress bar for better visualize the training)
pip3 install loggingandpip3 install tqdm -
Use the requirements.txt:
a) Recommend to use a virtualenv to install all files in the requirements.txt file.
b) cd to the directory where requirements.txt is located.
c) activate your virtualenv.
d) run:
pip3 install -r requirements.txtin the terminal -
Run the DockerFile:
a) To get started, you will need to first install Docker on your machine
b) cd to the the directory where Dockerfile is located.
c) run:
docker build -t imageName:tagName ., note that you will name your own image and tagd) now you will have a new image with the imageName you define if you run
docker image ls
Please make sure your directories/files are satisfied the following structure before running the code.
.
├── data
│ ├── index
│ ├── processed_note_file
│ │ ├── p1,p2...p_n.txt
│ │ └── label.txt
├── results
│ └── models
├── mimic_csv
│ │ ├── NOTEEVENTS.csv
│ │ ├── ADMISSIONS.csv
│ │ └── PATIENTS.csv
├── pre-trained_embedding
│ └── w2v_mimic.txt
├── tensorboard_tsne
│ ├── tensorboard_log
│ ├── extract_patient_vector.py
│ ├── generate_tsen_data.py
│ └── tensorboard_tsne.py
├── Embedding.py
├── HP.py
├── train.py
├── README.md
├── mimic_preprocessing.py
├── preprocessing_utilities.py
└── utility.pyTo get the result in the paper, run throughout the following steps:
-
Data Preprocessing
a. Download the mimic note data (.csv) and admission data (.csv)
b. Preprocessing and extracting the mimic note data specific for the project
python3 mimic_preprocessing.pyThis will call
preprocess_utilities.pyas the utility function.The result will generate a pandas dataframe with each patient information one line.
-
Prepare the embedding
a. Download the pre-trained word embedding
b. The function of
load_x_data_for_cnninutility.pyis used to load embedding for the model -
Train the model
a. The main model is designed in the function of
CNN_modelinutility.pyb. Train the model, evaluate and get the report score AUC
python3 train.py -
Generate tsne visualization
a. Extract the trained patient vector
python3 extract_patient_vector.pyb. Generate tsne data, feature and label
python3 generate_tsne_data.pyc. Tensorboard: then open localhost:6006 in a browser
tensorboard --logdir=/path/to/log --port=6006
The hyperparameters for deep learning and other arguments used in the code are all documented in HP.py. For the future development of this repo, we will introduce argparse for inputting parameters.
The performance is evaluated on AUROC score.
-
Performance on single task learning on four tasks:
Task AUC on single task In-hospital 93.90 30-day 93.05 60-day 92.32 1-year 90.39 -
Performance on multi-task models and report AUC on three related tasks:
Multi-task models In-hospital 30-day 1-year 3-task model 94.57 93.24 89.58 5-task model 94.07 93.07 90.56 20-task model 93.41 92.35 90.59 -
Apply the extracted patient vectors generated from 3-task, 5-task, 20-task models on target task of 60-day prediction:
AUC on 60-day mortality task 3-task patient vector 91.97 5-task patient vector 92.42 20-task patient vector 92.12 -
Performance of 3-task model(30-day, 1-year, 6-day LOS) on each related task:
Multi-task model 30-day mortality 1-year mortality 6-day LOS 3-task (30-day mortality,1-year mortality,6-day LOS) model 92.91 90.85 88.61 -
Apply the extracted patient vectors generated from 3-task(30-day, 1-year, 6-day LOS) on target task of 14-day LOS:
AUC on 14-day LOS task single task model 89.00 3-task (30-day, 1-year mortality, 6-day LOS) model 90.39 3-task (in-hospital, 30-day, 1-year mortality) model 74.71 -
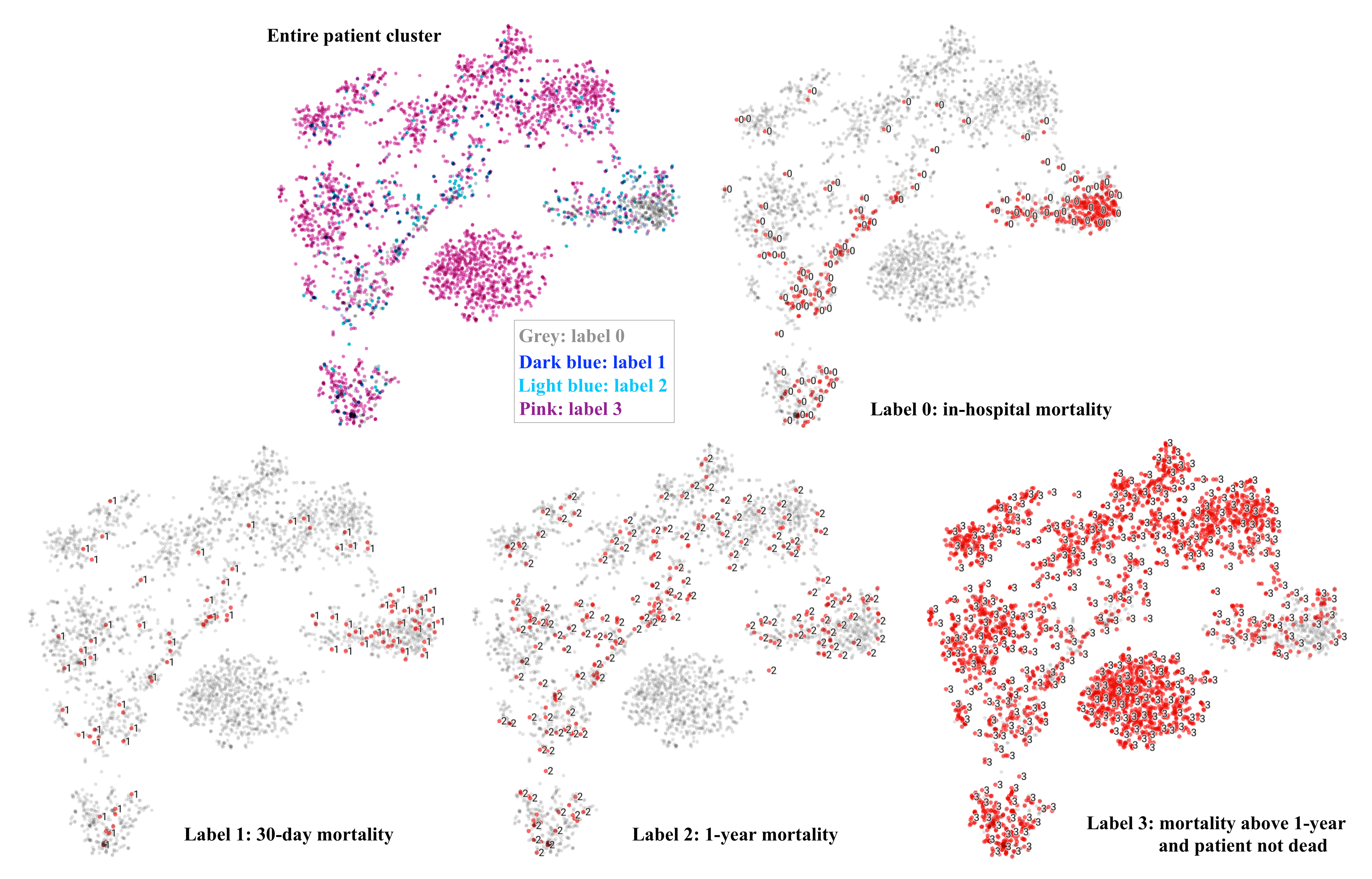
t-SNE on extracted 50-d patient vectors
- Si, Yuqi, and Kirk Roberts. "Deep Patient Representation of Clinical Notes via Multi-Task Learning for Mortality Prediction." AMIA Summits on Translational Science Proceedings 2019 (2019): 779.
@article{si2019deep,
title={Deep Patient Representation of Clinical Notes via Multi-Task Learning for Mortality Prediction},
author={Si, Yuqi and Roberts, Kirk},
journal={AMIA Summits on Translational Science Proceedings},
volume={2019},
pages={779},
year={2019},
publisher={American Medical Informatics Association}
}
Please contact Yuqi Si (yuqi.si@uth.tmc.edu) with questions and comments associated with the code.