A statistical tool to detect differential alternative splicing events using single-cell RNA-seq
For optimal performance, we recommend a HPC with 20+ cores
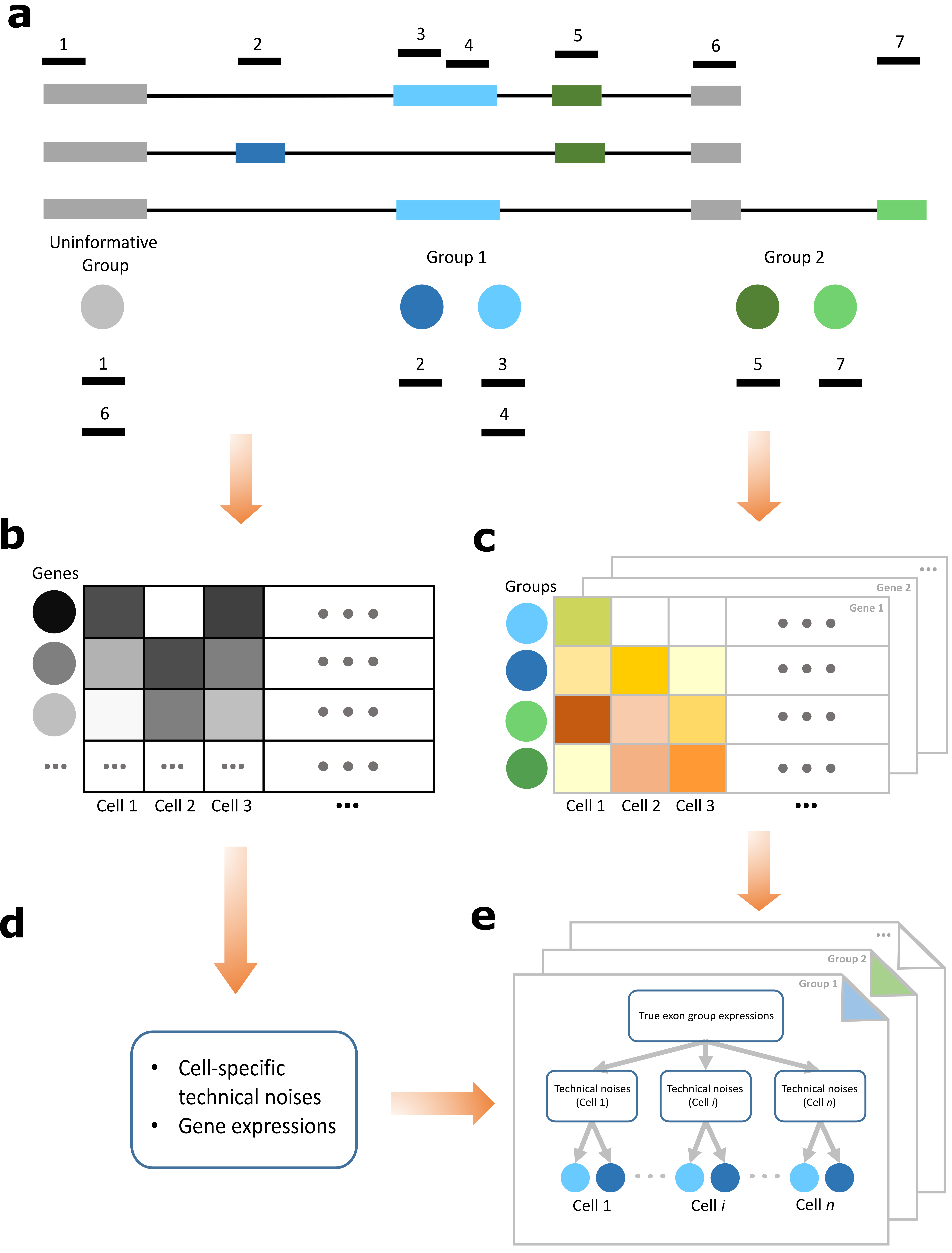
The input of SCATS is single-cell RNA-seq read data in BAM format together with a refrence isoform annotation file.
Please refer to Installation for how to install SCATS.
Please refere to Usage for how to use SCATS.
If you have any questions/issues/bugs, please post them on GitHub. They would also be helpful to other users.