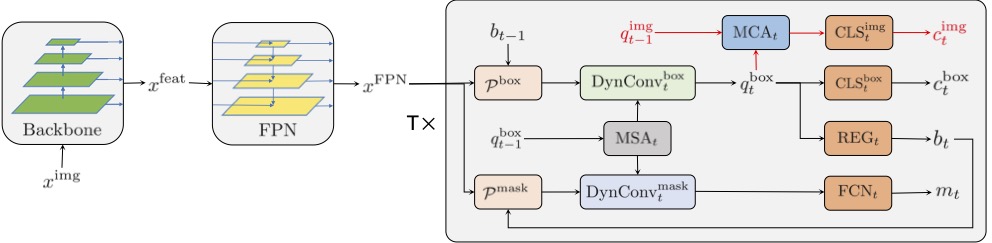
Query2: Query over Queries for Improving Gastrointestinal Stromal Tumour Detection in Endoscopic Ultrasound
More code will be released soon. I'm happy to provide full dataset and answer any Issues about this work. Please let me know if you need any help.
the data is shared under CC-by-NC-SA 4.0 license
Detail evaluation on each splits refers to 'work_dirs/queryglob_usdanno514roi_B_2_7_split<n>/test_log_queryglob_usdanno514roi_B_2_7_split<n>_sor.txt'
| Split_0 | Split_1 | Split_2 | Split_3 | Split_4 | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LMYM | GIST | LMYM | GIST | LMYM | GIST | LMYM | GIST | LMYM | GIST | ||
| T | 45 | 48 | 49 | 52 | 48 | 54 | 51 | 43 | 48 | 51 | 489 |
| F | 7 | 6 | 2 | 0 | 1 | 0 | 0 | 9 | 0 | 0 | 25 |
| MISS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acc | 87.74% | 98.06% | 99.03% | 91.26% | 100.00% | 95.14% | |||||
| Sen | 88.89% | 100.00% | 100.00% | 82.69% | 100.00% | 94.30% | |||||
| Spe | 86.54% | 96.08% | 97.96% | 100.00% | 100.00% | 96.02% |
training and test log: 'work_dirs'
@article{he2022query2,
title={Query2: Query over queries for improving gastrointestinal stromal tumour detection in an endoscopic ultrasound},
author={He, Qi and Bano, Sophia and Liu, Jing and Liu, Wentian and Stoyanov, Danail and Zuo, Siyang},
journal={Computers in Biology and Medicine},
pages={106424},
year={2022},
publisher={Elsevier}
}
- In file 'mmdet/datasets/anatomy.py', '_eval_global()' denotes the inference with SOR, while '_eval_vanilla' denotes inference without SOR.
- Code files for query^2 is located at 'mmdet/models/detectors/queryglob.py'. (and don't be confused if you see 'mmdet/models/detectors/query2.py', this is the other model we have tried but not publised)