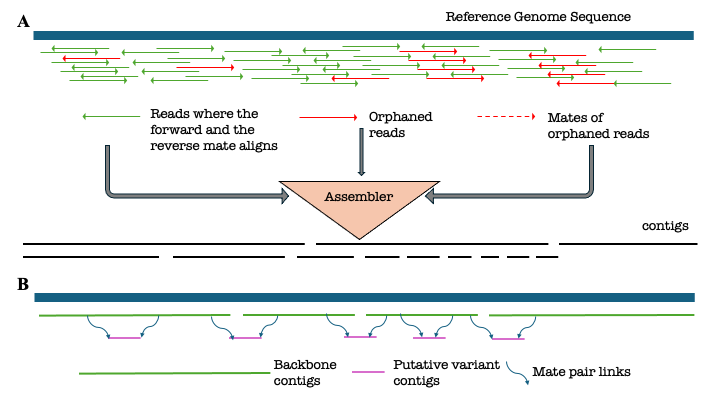
In this repository we maintain all the workflows to peform a reference reliant assembly and scaffolding of the hotspring metagenomes. The overall workflow can be summarized using this schematic diagram.
Snakefile.Reassemble_Reads.smk- Snakemake workflow to perform reference reliant assembly and scaffolding of the hotspring metagenomes. The config files associated with the workflow is available in theScripts/Synechococcus_Paper/configs/config.jsonSnakefile.Cluster.smk- Snakemake workflow to cluster the putative-variants identified by the previous step and annotates the variants using EggNOG. The config files associated with the workflow is available in theScripts/Synechococcus_Paper/configs/config_cluster.jsonSnakefile.Summarize.smk- Snakemake workflow to summarize the results from the previous steps and produces data files required for generating our figures/plots. The config files associated with the workflow is available in theScripts/Synechococcus_Paper/configs/config_summarize.json
This is a work in progress and reach out to hsmurali@umd.edu in case of any questions.