The discovery of antibiotics was an important milestone in advancement of modern medical science and has become an important aspect of healthcare, veterinary and agricultural industries. However, antimicrobial resistance, where the bacteria evolve rapidly to defeat drugs designed to kill them, is on the rise. Bacteriophages, viruses that infect and kill bacteria, are increasingly being explored as alternatives or complements to antibiotics. Phages have several advantages, including but not limited to, their host specificity, allowing them to target one organism rather than wiping out entire microbial communities, their ability to self replicate, and their ability to degrade biofilms which also reduce the efficacy of antibiotics. The effectiveness of phage therapy critically relies on the availability of a collection of phages able to infect the pathogens being targeted. Despite recent, substantial progress, the discovery of novel phages remains highly laborious.
Whole metagenomic shotgun sequencing datasets of mixed microbial community samples can be utilized to identify previously uncharacterized phages; it is estimated that up to 17% of the microbial DNA recovered from human stool samples comes from phage and a recent study of 70 animal microbiomes resulted in discovery of over 2,500 circular viral genomes. While sequencing data could substantially expand the number of phages available for phage therapy applications, identifying novel phage in these datasets is computationally difficult. Unlike prokaryotes and eukaryotes, which contain the phylogenetically informative 16S and 18S rRNA marker genes, respectively, viruses lack a single universal marker gene. Furthermore, phage genomes are significantly smaller than bacterial or fungal genomes, limiting their yield in whole metagenomic shotgun sequencing data. Existing bioinformatics tools for detecting phage from complex, fragmented metagenomes are optimized for longer contigs greater than 2 kb and often rely on homology to known viral references. Because it is difficult to isolate and characterize phage, these viral references are often limited and sparse.
In this study, we show that a subset of structural variants discovered in assembly graphs contain phage-like genomic elements. We applied MetaCarvel1, a tool developed in our lab for de novo scaffolding and reference independent variant discovery in metagenomic shotgun sequencing datasets, to 208 stool samples from the Human Microbiome Project (HMP)2. MetaCarvel constructs directed assembly graphs, where nodes represent unitigs, sequences that can be unambiguously reconstructed from the sequencing reads, while the edges indicate that the unitigs are adjacent in one or more organisms in the sample. The adjacency information is inferred either through sequence overlap information or through paired-end read information.
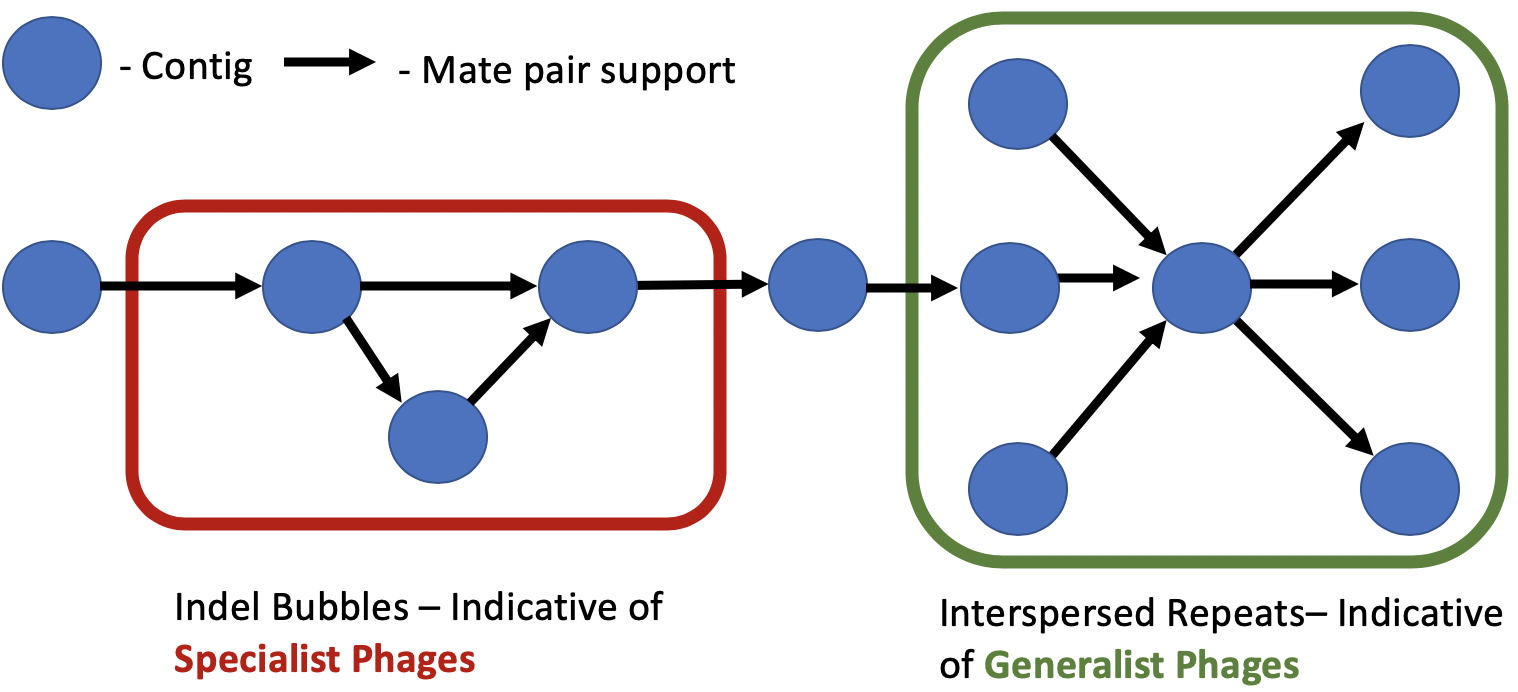
We observed contigs with phage elements in insertion-deletion (indel bubbles) and interspersed repeat variants detected by MetaCarvel. An indel bubble occurs when one path through the assembly graph contains the source and sink contigs, while other paths contain additional contigs that could indicate insertion or deletion events in a genome. Interspersed repeats manifest as high centrality nodes consisting of a single contig flanked by multiple different contigs. We believe that indel events could represent host-specific phage that integrate into certain genomes of a single species or strain, while interspersed repeats could represent phage genomes that are shared by many host species.
We aligned the contigs identified from predicted structural variants to crAssphage, a 97kb circular phage that was computationally identified and is prevalent in human fecal samples4. A third of the contigs aligning to crAssphage were found in the predicted interspersed repeats and 18% of these contigs were found in indel bubbles predicted by MetaCarvel. This suggests that MetaCarvel structural variants could be used to identify potentially novel phage. In the adjoining figure we show a crAssphage element found in an interspersed repeat,
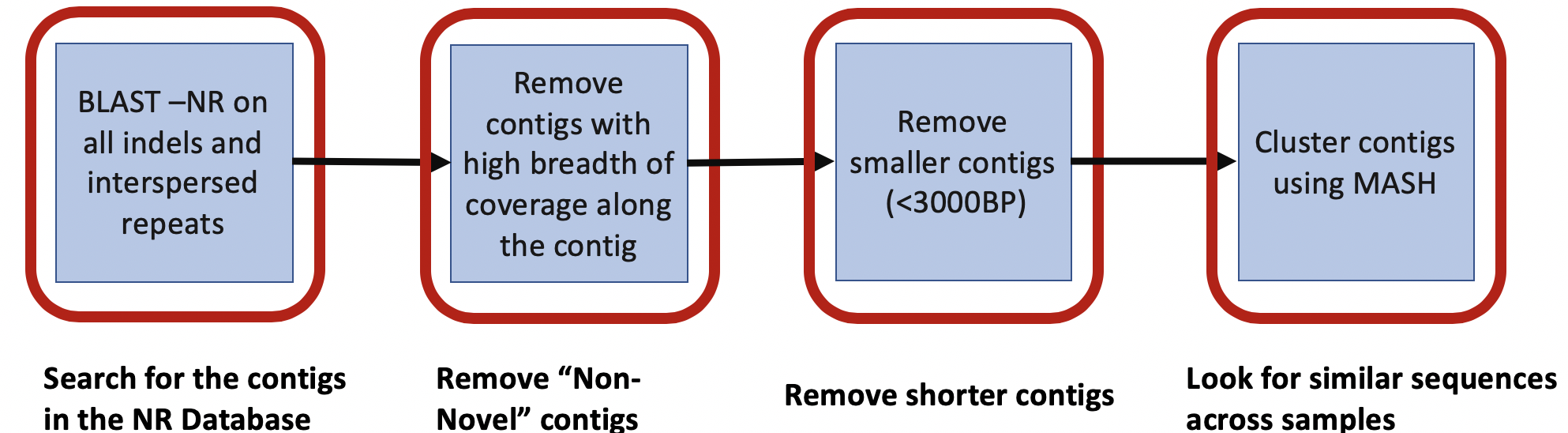
To search for novel phages, we collected contigs belonging to interspersed repeats within the 208 HMP stool samples. We then aligned these contigs against the NCBI non-redundant(NT) database and limited our focus to 104156 contigs greater than 2.5kbp that had no confident matches, and thus are potentially novel microbes. We used minimap2 to perform an all-vs-all alignment of the contigs and identified clusters of similar genomic content. The schematic pipeline is described below:
To identify sequences of interest, we run the following pipeline to preprocess.
python src/Preprocess.py -h
usage: Preprocess.py [-h] -i INPUT -l CONTIG_LIST -o OUTPUT_DIRECTORY
[-c BREADTH_COVERAGE_CUTOFF] [-len MIN_LEN]
[-t NUM_THREADS] [-pre PREFIX]
PIRATE: Phage Identification fRom Assembly-graph varianT Elements.This is the
pieline to filter potential repeat contigs of interest that is likely to
contain novel phage-elements. To that end, we align all contigs identified as
interspersed repeats against the non-redundant -nt database and retain only
poorly covered contigs for further downstream tasks.
optional arguments:
-h, --help show this help message and exit
-i INPUT, --input INPUT
A fasta file of assembled contigs
-l CONTIG_LIST, --contig_list CONTIG_LIST
A test file of contigs to be considered for putative
phage discovery
-o OUTPUT_DIRECTORY, --output_directory OUTPUT_DIRECTORY
Directory to write outputs to.
-c BREADTH_COVERAGE_CUTOFF, --breadth_coverage_cutoff BREADTH_COVERAGE_CUTOFF
breadth of coverage to filter contigs. (default = 50)
-len MIN_LEN, --min_len MIN_LEN
minimum length of contigs to considered to retain for
further analyses. (default = 3000)
-t NUM_THREADS, --num_threads NUM_THREADS
Number of threads. (default = 8)
-pre PREFIX, --prefix PREFIX
Prefix for output files (default = PIRATE.preprocess)
To cluster sequences using we run the following pipeline,
python src/PIRATE.py
usage: PIRATE.py [-h] -i INPUT [-d PERCENT_IDENTITY_CUTOFF]
[-c CONTAINMENT_CUTOFF] [-o OVERHANG] [-t NUM_THREADS] -out
OUTPUT_DIRECTORY [-pre PREFIX]
PIRATE: Phage Identification fRom Assembly-graph varianT Elements.PIRATE is a
pipeline for identifying phage elements from assembly graph variants such as
interspersed repeats and indel bubblesWe assume MetaCarvel was run on
metagenomic assemblies and variants were called out. After filtering out
contigs found in databases with sufficient coverage, we run PIRATE.
optional arguments:
-h, --help show this help message and exit
-i INPUT, --input INPUT
A fasta file of sequences to cluster
-d PERCENT_IDENTITY_CUTOFF, --percent_identity_cutoff PERCENT_IDENTITY_CUTOFF
percent identity to keep contigs within the same
cluster.(default = 95)
-c CONTAINMENT_CUTOFF, --containment_cutoff CONTAINMENT_CUTOFF
Fraction of the query contig to be covered by the
centroid contig to retain within the cluster.(default
= 0.80)
-o OVERHANG, --overhang OVERHANG
Length of the query contig overhanging with respect to
the cluster centorid. (default = 100bp)
-t NUM_THREADS, --num_threads NUM_THREADS
Number of threads. (default = 8)
-out OUTPUT_DIRECTORY, --output_directory OUTPUT_DIRECTORY
Location to write the outputs to
-pre PREFIX, --prefix PREFIX
Prefix for output files. (default = PIRATE)
We predicted genes using prodigal and annotated them using EggNOG.
bash src/Annotate.sh <fasta file of input sequences(seqs.fa)> <output directory> <prefix>
- Ghurye, J., Treangen, T., Fedarko, M., Hervey, W. J., 4th & Pop, M. MetaCarvel: linking assembly graph motifs to biological variants. Genome Biol. 20, 174 (2019).
- Consortium, T. H. M. P. & The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature vol. 486 207–214 (2012).
- Heng Li. Minimap2: pairwise alignment for nucleotide sequences, Bioinformatics, 34, 18, 3094–3100 (2018).
- Edwards, R. A. et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat Microbiol 4, 1727–1736 (2019).