Equivalent Graph Neural Network-based Accurate and Ultra-fast Virtual Screening of Small Molecules Targeting miRNA-Protein Complex
If you find it useful, please cite:
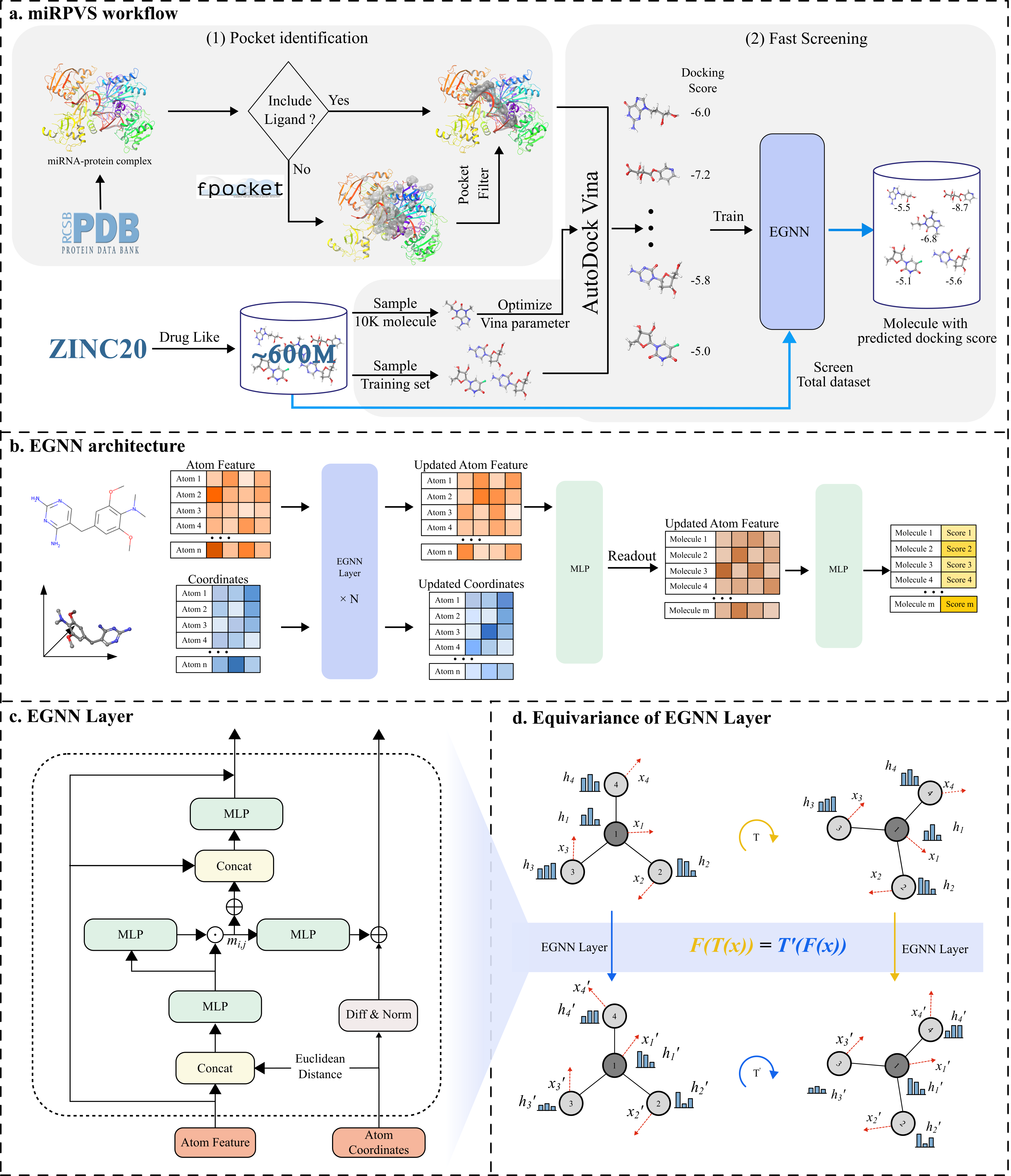
Equivalent Graph Neural Network-based Virtual Screening of Ultra-large chemical libraries Targeting miRNA-protein complex Huabei Wang; Zhimin Zhang; Guangyang Zhang, Ming Wen* and Hongmei Lu*. Will Published in: DOI:
autodock vina
python
The package development version is tested on Linux: Ubuntu 22.04 operating systems.
Dependencies for SMTarRNA:
pytorch
pyg
rdkit=2022.09.1
git clone https://github.com/huabei/miRPVS.git
you can install the env via yaml file
cd miRPVS
conda env create -f requirements.yaml
this project use ashleve/lightning-hydra-template as the base project.
This template is suitable for multi-platform operation, please note that the config/local is configured specifically for different platforms.
You just need to configure your own hyperparameters in config/experiment and then run:
python src/train.py experiment=exp_nameThe configuration used for this job is also stored in the config/experiment directory and can be used directly.
The config/eval.yaml file needs to be configured with your data locations, model parameter paths, etc. And run:
python src/eval.pyThe config/predict.yaml file needs to be configured with your data locations, model parameter paths, etc. And run:
python src/predict.py