| title | output | |||
|---|---|---|---|---|
Lab 12 - Create a website |
|
library(data.table)
library(tidyverse)
library(dplyr)
library(plotly)
library(DT)
library(knitr)
# INITALIZE CODE CHUNK OPTIONS
opts_chunk$set(
warning = FALSE,
message = FALSE,
eval=TRUE,
echo = TRUE,
cache = FALSE,
fig.width = 7,
fig.align = 'center',
fig.asp = 0.618,
out.width = "700px",
class.source = "code-r")
.code-r { /* Code block */
font-size: 15px;
}
.code-r-small { /* Code block */
font-size: 10px;
}
The objective of today's lab is to create a website using the rmarkdown package and deploy it using GitHub pages.
We will include interactive visualizations (e.g. plot_ly() output) on the website.
For this lab, we have created some initial content for you to work with for an example website: https://github.com/JSC370/jsc370-2022/tree/main/labs/lab12
Your objective will be to create your own personal project website using this as a reference.
Building websites uses the same reproducible workflow you can use for your analyses and collaborations. It is very iterative. You can do it all from RStudio, with a combination of clicking or typing commands as you feel comfortable.
There are two main steps for creating a personal website that will be hosted on GitHub:
I. Local setup
II. GitHub setup
The basic workflow is as follows:
- Create a project directory and an R Project file (
.Rproj) - Create a
_site.ymlandindex.Rmdfile in your new directory - Add additional page content if desired through other
.Rmdfiles - Edit these files to create content and manage layout (and knit to view)
- Add a style sheet (CSS) if desired
- Build website
- Build tab > Build Website or
- in the console:
rmarkdown::render_site()This creates the output:index.html
Look at the contents of the lab 12 repo. What files does it contain?
- Take a few moments to compare the contents of this repository to the final website (from a previous student) at https://abigailhorn.github.io/PM566-final-project
Create and go to the directory you want to create your website in, e.g. "my-website", and initialize git.
In command line:
mkdir my-website
cd my-website
git initRecall from lecture that the minimum requirements for an R Markdown website are:
index.Rmd: contains the content for the website homepage_site.yml: contains metadata for the website
Create these essential files, as well as a README.md, add all to git queue, and commit to your website repository.
Note: Use the touch command from command line to initialize empty files:
echo My JSC370 Final Project Website > README.md
touch _site.yml
touch index.Rmd
git add --all
git commit -m "initalizing repository"Create an R Project file using RStudio IDE:
Go to RStudio IDE > File > New Project > Existing Directory
The R Project is useful because RStudio will recognize your project as a website, and provide appropriate build tools.
Note: After creating the R Project and initial files, you may need to close the project and reopen it before R will recognize it as a website and show the appropriate build tools.
Edit the _site.yml file to include the metadata, layout, and theme you want for your website.
First let's take a look at a basic example of a _site.yml file for a website with one page:
name: "my-website"
navbar:
title: "My Website"
left:
- text: "Home"
href: index.htmlThis is the minimum you need to include in your _site.yml.
Now let's take a look at the _site.yml from the website repository you downloaded into "lab-12". It looks like this:
name: "my-website"
output_dir: "."
navbar:
title: "JSC370 Final Project"
left:
- text: "Home"
href: index.html
right:
- icon: fa-github fa-lg
href: https://github.com/JSC370/jsc370-2022/
- text: "JSC370 Home"
href: https://jsc370.github.io/jsc370-2022/
output:
html_document:
theme: cosmo
include:
after_body: footer.html
css: styles.cssInspecting this, and the output on the completed website at https://abigailhorn.github.io/PM566-final-project, how do you add links to internal webpages? How do you add links to external websites? How do you add icons?
Note: recall that the output_dir field indicates which directory to copy site content into ("_site" is the default if none is specified). It can be "." to keep all content within the root website directory alongside the source code.
Note: Preview themes here and play around with different options. Themes are easy to change even after you have added content.
Now your task is to create a YAML for your website that includes only the essential components for your website. Either copy the content of the simple _site.yml into your own _site.yml file in your website directory my-website, or replicate it yourself line by line.
Edit and create .Rmd files that contain your website content, which will produce the html pages of your website when you knit them.
For example, the index.Rmd could look like this:
---
title: "JSC370 Final Project"
author: "Your Name"
output:
html_document:
toc: TRUE
toc_float: TRUE
---
This is my JSC370 Final Project website.
Recall that the toc specifies whether there is a table of contents, and toc_float provides the option to float the table of contents to the left of the main document content. The floating table of contents will always be visible even when the document is scrolled. There are other options for how to display the toc in R Markdown HTML output which you can read about here.
After you are done with your index.Rmd file, knit it to check the output. Either click the Knit option in the toolbar or in the console type rmarkdown::render_site("index.Rmd"). This will render the output into a file index.html which you can check out by opening the file in your directory:
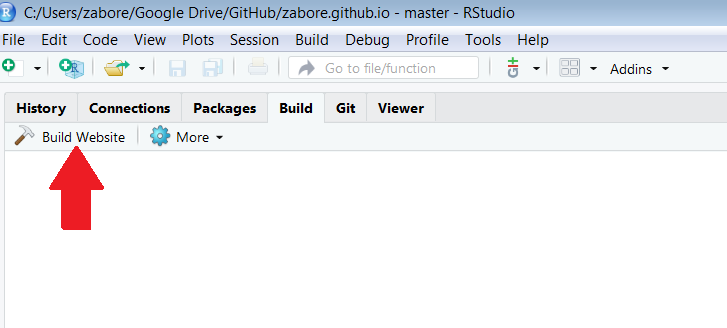
Now we have the content and layout setup, we can build the website! This can be done in two ways:
- Build tab > Build Website
- in the console:
rmarkdown::render_site()
rmarkdown has created all the additional files you need for your website. Check them out in your directory. Most importantly, the index.html file provides with a preview of the site, which you can look at in a browser as above:
- Create project on GitHub
- Initialize project on Git
- Push project files to the GitHub repository for your project
- Deploy the website by enabling GitHub pages for the repository
Create an online (remote) repository for your project using GitHub
In command line:
Add the remote using git remote add
git remote add origin https://github.com/YOUR_GITHUB_NAME/YOUR_PROJECT_NAME.gitOptionally, use the commands git status and git remote -v to check out the status.
Push the changes to the remote using git push
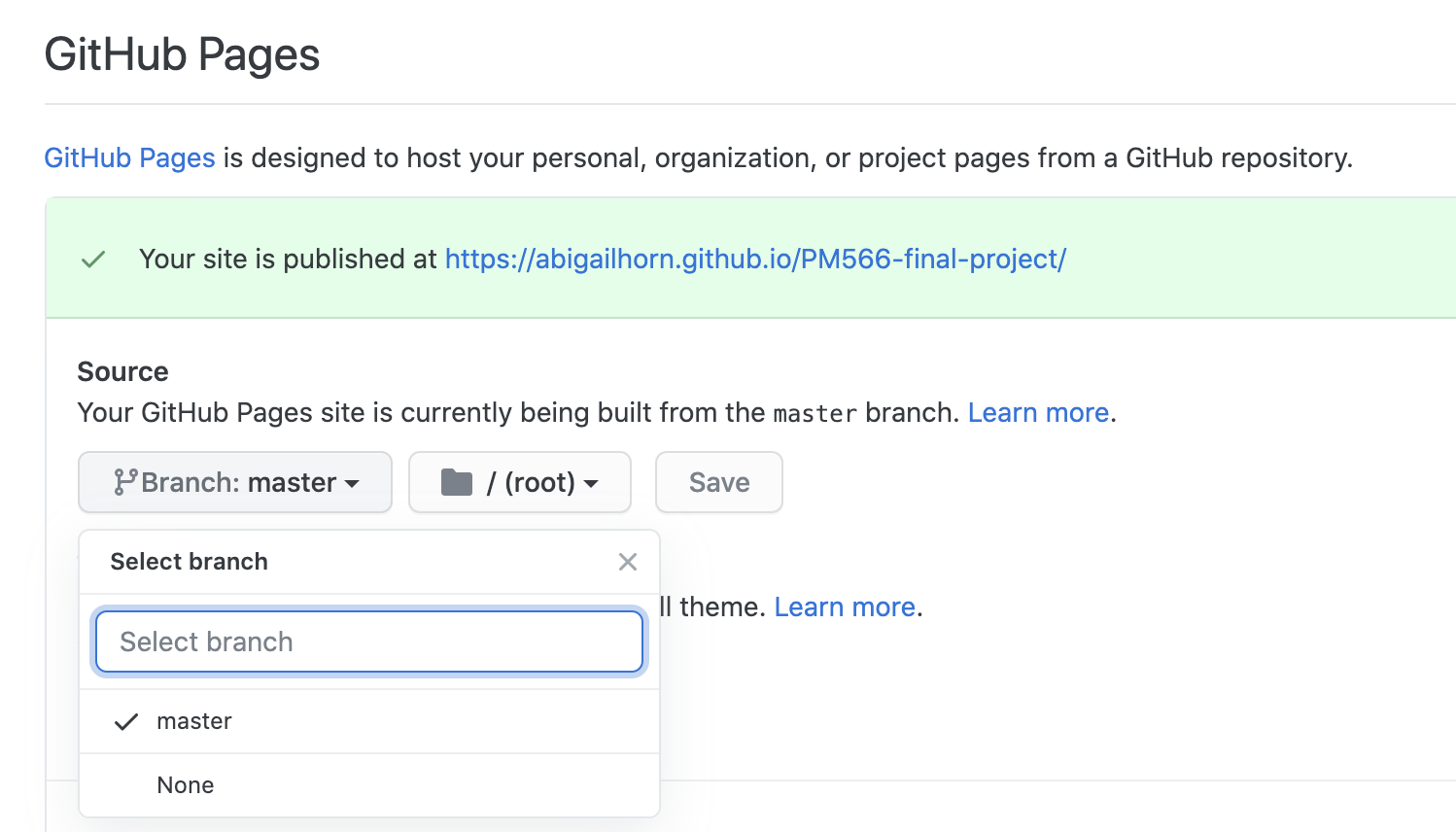
git push -u origin masterEnable GitHub pages for the repository by going to the repository's Settings > GitHub Pages, where you'll select the "master branch" folder and hit Save:
It's live! Go to the website at www.YOUR_GH_NAME.github.io/YOUR_PROJECT_NAME/ (the website should appear to you when you click the appropriate setting in GitHub Pages)
Your task here is to create 2 interactive visuals, using plotly, leaflet, DT, or anything else you have explored, and post them on your website at index.Rmd.
First you can source any necessary code, meaning run it. For example, let's use the COVID-19 data from the NYT we explored in week 11. In the lab repository, we have provided the code process_covid_data.R which goes through the first steps we carried out in the lab of downloading and processing the data. To source this code:
fn <- "https://raw.githubusercontent.com/JSC370/jsc370-2022/main/labs/lab12/process_COVID_data.R"
if (!file.exists("process_COVID_data.R"))
download.file(fn, destfile = "process_COVID_data.R")
source("process_COVID_data.R")
Recall that echo=FALSE means the code itself will not appear in the HTML output. You can also set this globally in opts_chunk$set(echo=FALSE) (see above for this lab)
Then you can add some code chunks to create the interactive visuals you want to include. I will add some code to create a couple of the plotly figures we created in lab. I am naming each plot but not outputting them here, because I will want to do that in independent code chunks as we will see in the next step.
Note: Code chunks cannot have the same name, so if you do name them (like this one: plot1), you will need to be sure to give each an independent name.
p1_scatter <- cv_states_today %>%
plot_ly(x = ~pop_density, y = ~deathsper100k,
type = 'scatter', mode = 'markers', color = ~state,
size = ~population, sizes = c(5, 70), marker = list(sizemode='diameter', opacity=0.5),
hoverinfo = 'text',
text = ~paste( paste(state, ":", sep=""), paste(" Cases per 100k: ", per100k, sep="") , paste(" Deaths per 100k: ",
deathsper100k, sep=""), sep = "<br>")) %>%
layout(title = "Population-normalized COVID-19 deaths vs. population density",
yaxis = list(title = "Deaths per 100k"), xaxis = list(title = "Population Density"),
hovermode = "compare")
# filter out "District of Columbia"
cv_states_today_scatter <- cv_states_today %>% filter(state!="District of Columbia")
p2_scatter <- cv_states_today_scatter %>%
plot_ly(x = ~pop_density, y = ~deathsper100k,
type = 'scatter', mode = 'markers', color = ~state,
size = ~population, sizes = c(5, 70), marker = list(sizemode='diameter', opacity=0.5),
hoverinfo = 'text',
text = ~paste( paste(state, ":", sep=""), paste(" Cases per 100k: ", per100k, sep="") , paste(" Deaths per 100k: ",
deathsper100k, sep=""), sep = "<br>")) %>%
layout(title = "Population-normalized COVID-19 deaths vs. population density",
yaxis = list(title = "Deaths per 100k"), xaxis = list(title = "Population Density"),
hovermode = "compare")
Now, please create 2 figures of your own, either using the code from last week's lab, or creating new figures based on the data created by the process_COVID_data.R code.
Create tabs to display each figure. We do that using the following R Markdown language:
## Showcasing plots {.tabset}
### Figure 1
`r ''````{r echo=FALSE}
p1_scatter
```
### Figure 2
`r ''````{r echo=FALSE}
p2_scatter
```
{-}
The output will look like this:
p1_scatter
p2_scatter
Knit the page index.Rmd to check the output. It may take a bit longer now that we're also processing the data from the NYT. (Recall from lecture we can do that once per session by inputting the global option opts_chunk$set(cache=TRUE)).
You've now made some edits to your website. To get the updates onto the live webpage, you need to re-render the site to create the HTML output from your .Rmd file edits, and push the updates to the remote GitHub repository:
-
In the R console:
rmarkdown::render_site() -
Preview contents by looking at the
index.htmlfile in a browser -
Add and push changes to remote from your website project repository locally (e.g.
JSC370-final-project):
git add --all
git commit -m "interactive visuals"
git push -u origin masterPreview your changes online at your website! Note that it may take up to 10 minutes for the content to render.
Add the online link to your website in your README.md file, e.g.
This is my JSC370 Final Project website home. The website is online at https://github.com/meredithfranklin/my-website.
Then please submit your lab by creating a document with the link to your github site and upload to quercus.
This lab pulled from:
- Chapter 10.5: Websites in rmarkdown’s site generator in R Markdown: The Definitive Guide, Yihui Xie, J. J. Allaire, Garrett Grolemund
- Creating websites in R, Emily C. Zabor