SIREN: A Scalable Isotropic Recursive Column Multimodal Neural Architecture with Device State Recognition Use-Case
- Siren
- Table of Contents
- Introduction
- Installation
- Usage
- SIREN Colab Demo
- Hugging Face Space
- Data
- Results
- Classification
- Table: Sensitivity studies of number of columns on Pentostreda dataset.
- Table: Sensitivity studies of depth of blocks on Pentostreda dataset.
- Table: Performance comparison of various models and modalities on the Pentostreda dataset.
- Results of MOSI and MOSEI
- Depth-Point and Intermediate activation removal
- Fixing the model size
- Multimodal Datasets
- Interpretability
- Contributing
Effective monitoring of device conditions and potential damage is a critical task in various industries, including manufacturing, healthcare, and transportation. The collection of data from multiple sources poses a challenge in achieving accurate device state recognition due to its complexity and variability. To address this challenge, we propose the use of multimodal data fusion through the combination of modalities, utilizing the concentration phenomenon, to establish appropriate decision boundaries between device state regions. We introduce Siren, a novel supervised multimodal, isotropic neural architecture with patch embedding, which effectively describes device states.
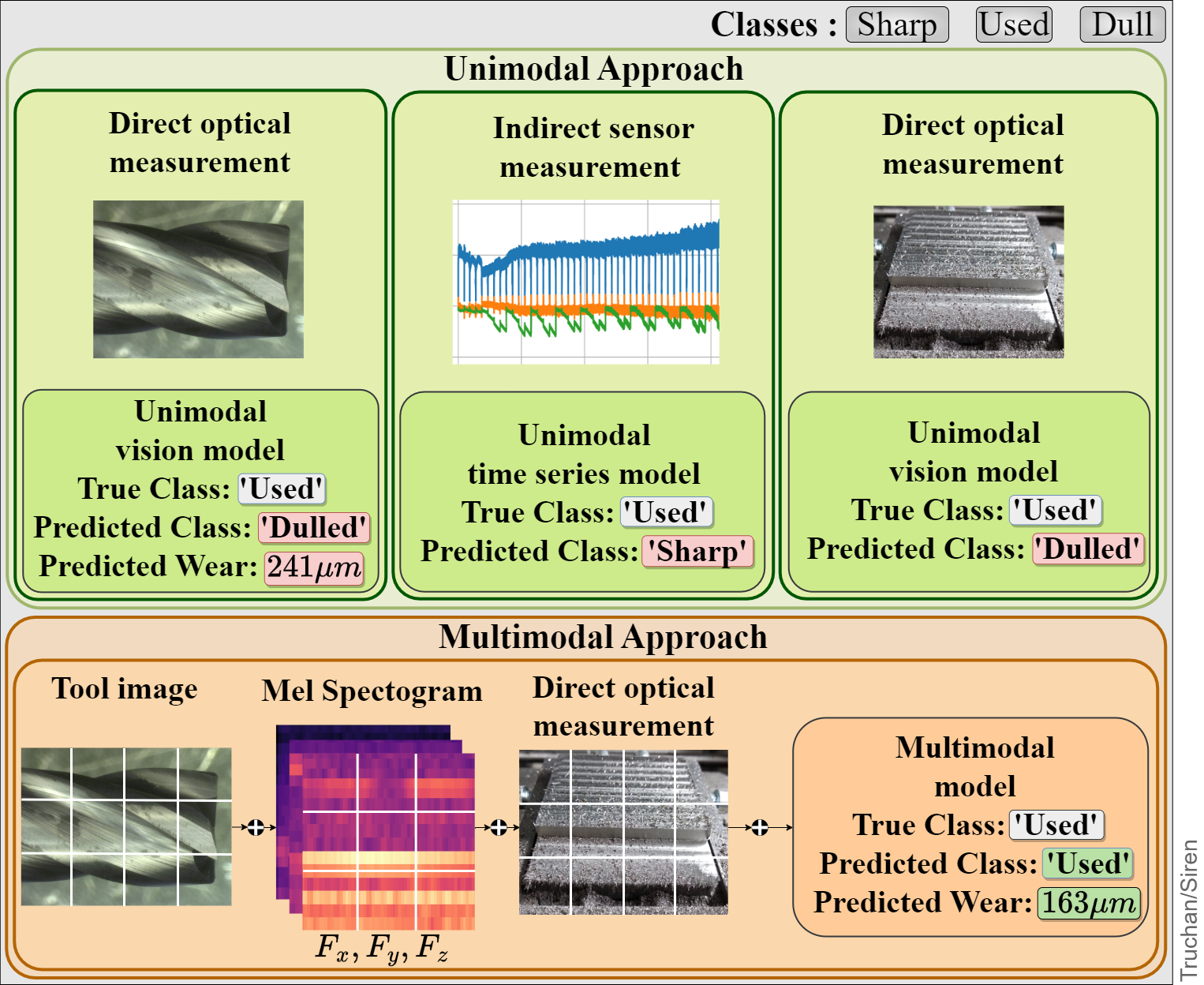
Figure: A sample instance from Pentostreda that shows the importance of leveraging multimodal data for device state prediction.
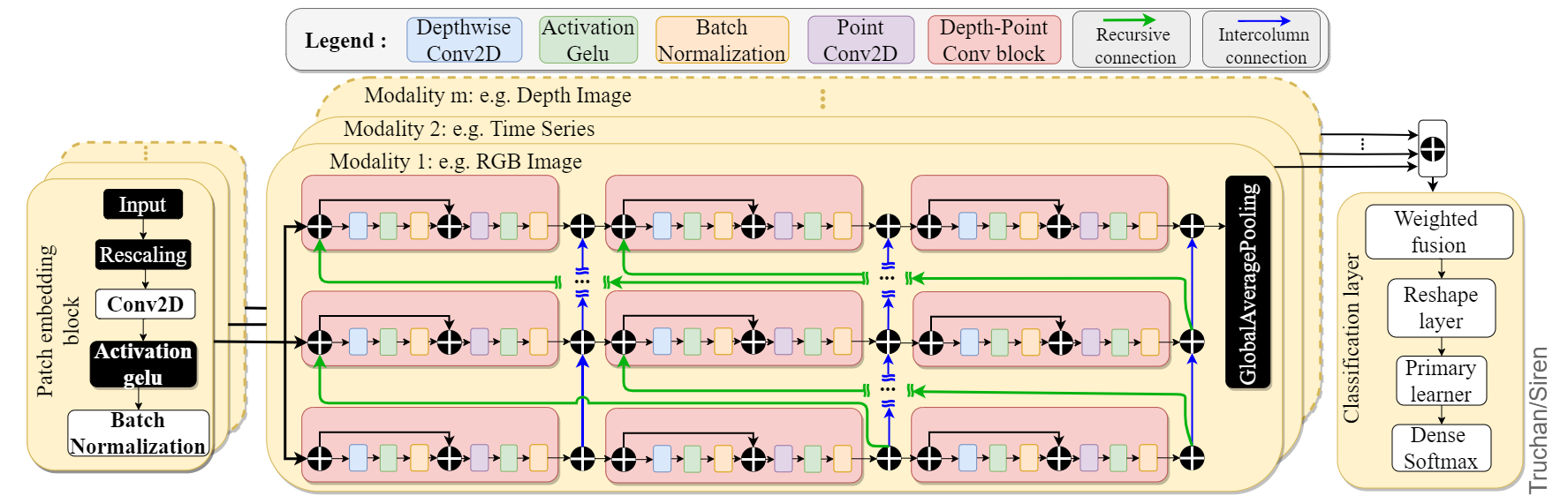
The Siren uses the linearly embedded patches of the grouped signals, applies the isotropic architecture for representation retrieval, and uses TempMixer recurrent structure to map their temporal dependencies
In section Data section we present the Pentostreda : the publicly available and accessible multimodal device state recognition dataset as a new benchmark for multimodal industrial device state recognition.
Note that this work requires Python version 3.9 or later.
Install the dependencies from the requirements.txt with:
pip install -r requirements.txt
If you want to run the model on your GPU make sure to follow the installation instructions from Tensorflow.
To run .ipynb files you have to install Jupyter Notebook/JupyterLab.
In order to clone the repository to your local machine use this command
git clone https://github.com/hubtru/Siren.gitTo use this project, follow these steps:
- Download the project files to your local machine.
- Choose the script that you want to use.
- One or multiple unimodal classification models:
- siren_base_tool.ipynb, train and save unimodal network for tool images.
- siren_base_spec.ipynb, train and save unimodal network for spectrogram images.
- siren_base_chip.ipynb, train and save unimodal network for chip images.
- One or multiple multimodal classification models (requires saved unimodal .h5 models):
- siren_base_multi_ts.ipynb, train and save multimodal network for tool and spectrogram images.
- siren_base_multi_sc.ipynb, train and save multimodal network for spectrogram and chip images.
- siren_base_multi_tc.ipynb, train and save multimodal network for tool and chip images.
- siren_base_multi_tsc.ipynb, train and save multimodal network for tool, spectrogram and chip images.
- One or multiple unimodal regression models:
- siren_reg_base_tool.ipynb, train and save unimodal network for tool images.
- siren_reg_base_spec.ipynb, train and save unimodal network for spectrogram images.
- siren_reg_base_chip.ipynb, train and save unimodal network for chip images.
- One or multiple multimodal regression models (requires saved .h5 unimodal regression models):
- siren_reg_base_multi_ts.ipynb, train and save multimodal network for tool and spectrogram images.
- siren_reg_base_multi_sc.ipynb, train and save multimodal network for spectrogram and chip images.
- siren_reg_base_multi_tc.ipynb, train and save multimodal network for tool and chip images.
- siren_reg_base_multi_tsc.ipynb, train and save multimodal network for tool, spectrogram and chip images.
- Update the paths section in each notebook if you want they differ from the recommended setup.
- Change the variables section if desired.
- Run the script
Those Notebooks will run fine with the structure of the git repository. If you want to change dataset paths check the Data Structures section.
The following table showcases the SIREN algorithm demonstrations for both unimodal and multimodal tasks, including their respective modalities, signals, tasks, and descriptions along with links to Colab notebooks.
Check out our demonstrator on Hugging Face Spaces.
| Description | Link | Task |
|---|---|---|
| Siren Demonstrator | Classification & Regression |
Pentostreda: Pento-modal Device State Recognition Dataset
Pentostreda is a five-modal data set collected from real-world milling processes over a span of seven weeks. It includes an RGB image of the milling tool, three-time series recordings of force signals, and an image that captures the metallic chips generated during the milling process. After every milling activity, an expert examined the tool's flank wear from a taken image to annotate the dataset. Depending on the observed tool wear, each instance was systematically categorized into one of three classes: new, usable, or dulled. As a result, we have created the multimodal dataset with labels, allowing us to solve the classification problem of the machine state.
- Data preview (~20 samples) is available in ./dataset
- Complete Pentostreda dataset (918MB) is available: Pentostreda (available soon)
- Complete Pentostreda.zip file (918MB) is available: Pentostreda.zip (available soon)
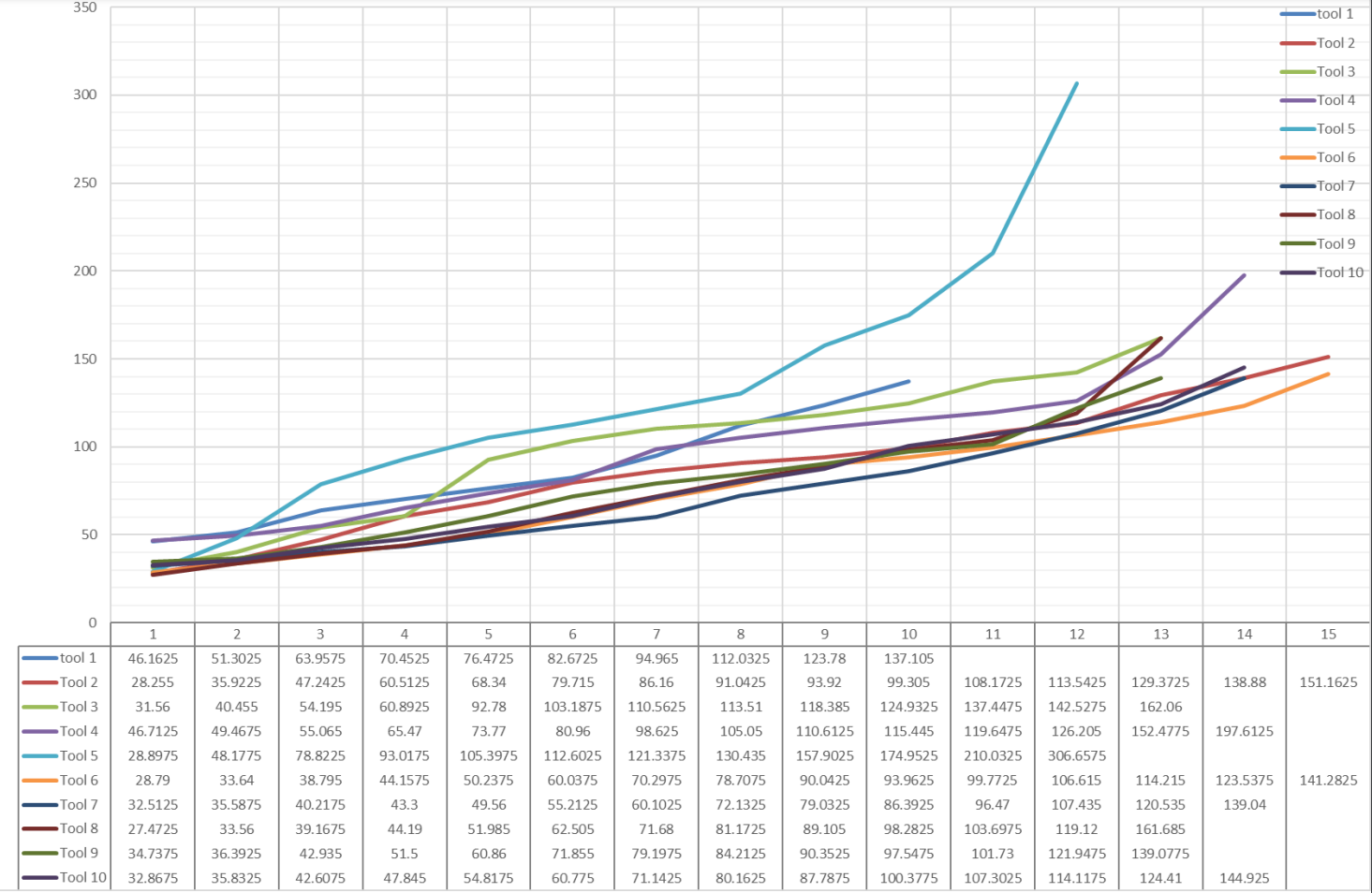
Figure: The flank tool wear measured with Keyence VHX- S15F profile measurement unit microscope for all ten tools. The values are presented in μm.
This section provides information about the data used in the project, including where it came from, how it was collected or generated, and any preprocessing or cleaning that was done.
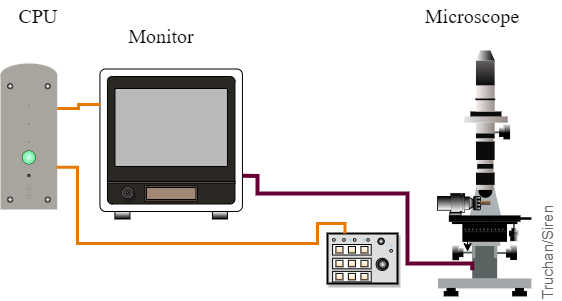
Figure: Measuring stage with the digital microscope and the industrial computer. |
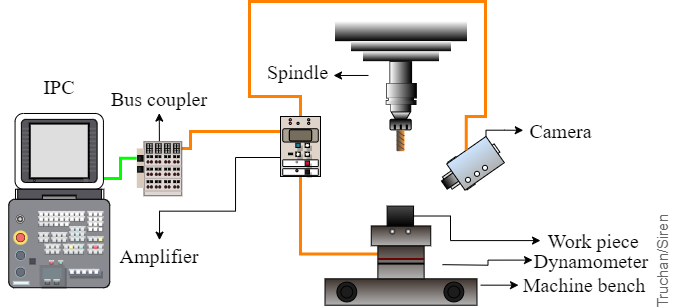
Figure: Processing stage with the signal recording diagram |
Figure The forces signals depicted on Figure Signal present 30 milling phases. After each phase, a picture of the tool was taken with an industrial microscope to determine its exact wear. A strong correlation between the tool wear and the force amplitudes can be observed, with the smallest amplitudes for the sharp tool, increasing with tool wear.
The summary of the Pentostreda data-set with classes samples of Tool 1.
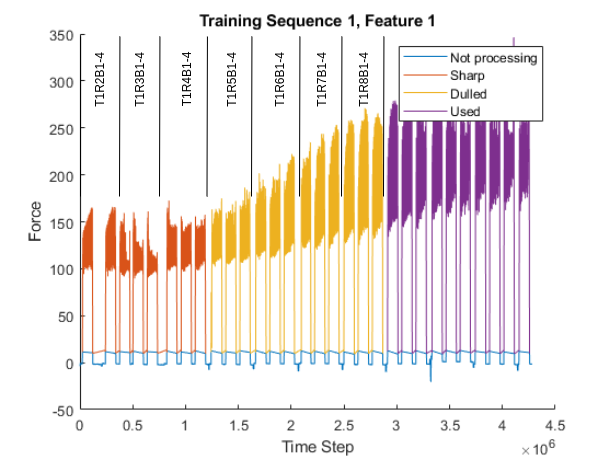
Figure: The summary of the **Pentostreda** data-set with classes samples of Tool 1.
.
For more visualisation, see: Forces-Visualisation
The folder contains the pictures of the flank wear, pictures of metal workpeace chips and the spectrograms of the forces in 3 axes (Fx, Fy, Fz).
dataset/
│
├── chip/
├── spec/x/
├── spec/y/
├── spec/z/
├── tool
│
├── labels.csv
├── labels_reg.csv
├── labels_sample.csv
└── labels_reg_sample.csvThis section provides a summary of the results of the project, including any performance metrics or visualizations that were produced. It should also include a discussion of the results and their implications.
| Class | Precision | Recall | F1-score |
|---|---|---|---|
| Sharp | 1.00 | 1.00 | 1.00 |
| Used | 1.00 | 0.95 | 0.97 |
| Dulled | 0.92 | 1.00 | 0.96 |
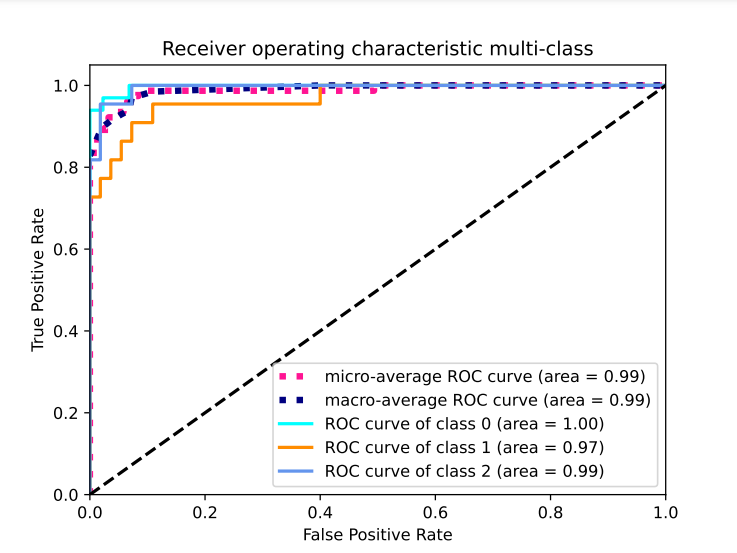
Figure: Receiver operating multi-class characteristic for SIREN hyper-band optimised multimodal (TSC: Tool, Sectrogram, Chip) model.
| #Column | Flops | Size (MB) | #Params | Img/sec | Accuracy (%) |
|---|---|---|---|---|---|
| 2 | 2711864 | 11 | 2,7M | 0s 17ms/step | 72.15 |
| 3 | 3575120 | 15 | 3,7M | 0s 20ms/step | 73.65 |
| 4 | 4438376 | 18 | 4,6M | 0s 24ms/step | 74.90 |
| 5 | 5301632 | 22 | 5,5M | 0s 25ms/step | 75.88 |
| 6 | 6164888 | 26 | 6,4M | 0s 28ms/step | 76.75 |
| 7 | 7028144 | 29 | 7,2M | 0s 32ms/step | 77.26 |
| 8 | 7891400 | 33 | 8,1M | 0s 32ms/step | 77.63 |
| 9 | 8754656 | 37 | 9,0M | 0s 39ms/step | 77.97 |
| 10 | 9617912 | 40 | 9,9M | 0s 44ms/step | 77.81 |
| Depth | Flops | Size (MB) | #Params | Img/sec | Accuracy (%) |
|---|---|---|---|---|---|
| 2 | 1988700 | 11 | 2.6M | 16ms/step | 71.63 |
| 3 | 2979266 | 15 | 3.6M | 21ms/step | 72.05 |
| 4 | 3631398 | 18 | 4.5M | 24ms/step | 74.90 |
| 5 | 4485996 | 22 | 5.6M | 53ms/step | 75.28 |
| 6 | 5527140 | 26 | 6.3M | 70ms/step | 76.43 |
| 7 | 6176247 | 29 | 7.3M | 82ms/step | 77.61 |
| 8 | 7038275 | 33 | 8.2M | 91ms/step | 77.84 |
| 9 | 8098056 | 37 | 8.9M | 87ms/step | 77.75 |
| 10 | 9583319 | 40 | 9.9M | 110ms/step | 77.53 |
| Model | Modality | Fusion-Type | Training | ms/img | Size(MB) | #Params | Flops | AUC | Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| Meta-Transformer | t | - | 1s 51ms/step | 78 | 993.3 | 86.6M | 85199412 | 0.85 | 57.1 (2) |
| Meta-Transformer | s | - | 1s 51ms/step | 78 | 993.3 | 86.6M | 85172200 | 0.84 | 60.4 (3) |
| Meta-Transformer | c | - | 1s 54ms/step | 78 | 993.3 | 86.6M | 84060374 | 0.85 | 63.9 (3) |
| Meta-Transformer | ts | ViT | 1s 78ms/step | 81 | 991.9 | 87.5M | 85819362 | 0.83 | 58.5 (3) |
| Meta-Transformer | tc | ViT | 1s 75ms/step | 81 | 991.9 | 87.5M | 86544254 | 0.85 | 65.3(4) |
| Meta-Transformer | sc | ViT | 1s 73ms/step | 81 | 991.9 | 87.5M | 83726432 | 0.84 | 61.1 (4) |
| Meta-Transformer | tsc | ViT | 1s 93ms/step | 83 | 1004.5 | 88.3M | 86367545 | 0.85 | 68.7 (4) |
| ViT | t | - | 3s 64ms/step | 36 | 417.4 | 36.4M | 36375641 | 0.87 | 56.5 (3) |
| ViT | s | - | 3s 69ms/step | 44 | 418.2 | 36.4M | 36473957 | 0.87 | 55.7 (4) |
| ViT | c | - | 3s 62ms/step | 35 | 417.4 | 36.5M | 36375641 | 0.89 | 66.3 (2) |
| ViT | ts | Concat | 5s 75ms/step | 69 | 836.1 | 72.9M | 72129427 | 0.88 | 59.2 (3) |
| ViT | tc | Concat | 5s 62ms/step | 59 | 835.4 | 72.8M | 71287741 | 0.89 | 66.4 (2) |
| ViT | sc | Concat | 5s 62ms/step | 61 | 836.1 | 72.9M | 73014364 | 0.89 | 67.6 (2) |
| ViT | tsc | Concat | 8s 12ms/step | 101 | 1252.1 | 109M | 109111829 | 0.90 | 68.8 (3) |
| MLP-Mixer | t | - | 1s 35ms/step | 26 | 25.7 | 2.2M | 2156650 | 0.87 | 57.1 (2) |
| MLP-Mixer | s | - | 1s 36ms/step | 36 | 30.3 | 2.6M | 2599088 | 0.88 | 61.3 (2) |
| MLP-Mixer | c | - | 1s 33ms/step | 25 | 26.8 | 2.3M | 2301206 | 0.87 | 55.4 (4) |
| MLP-Mixer | ts | Concat | 2s 76ms/step | 43 | 45.5 | 3.9M | 3895658 | 0.89 | 65.7 (2) |
| MLP-Mixer | tc | Concat | 2s 71ms/step | 46 | 40.9 | 3.5M | 3463465 | 0.88 | 57.6 (4) |
| MLP-Mixer | sc | Concat | 2s 78ms/step | 44 | 45.5 | 3.9M | 3826508 | 0.89 | 67.2 (3) |
| MLP-Mixer | tsc | Concat | 3s 67ms/step | 53 | 59.5 | 5.1M | 5086126 | 0.90 | 69.5 (2) |
| SIREN | t | - | 2s 47ms/step | 10 | 5.6 | 1.4M | 1382248 | 0.88 | 58.8 (1) |
| SIREN | s | - | 2s 53ms/step | 14 | 7.2 | 1.8M | 1775464 | 0.89 | 66.2 (1) |
| SIREN | c | - | 2s 44ms/step | 9 | 5.6 | 1.4M | 1382248 | 0.90 | 72.5 (1) |
| SIREN | ts | Concat | 4s 26ms/step | 19 | 12.8 | 3.2M | 3089991 | 0.90 | 72.9 (1) |
| SIREN | tc | Concat | 4s 25ms/step | 14 | 11.2 | 2.8M | 2696775 | 0.90 | 73.7 (1) |
| SIREN | sc | Concat | 4s 30ms/step | 16 | 12.8 | 3.2M | 3089991 | 0.90 | 73.3 (1) |
| SIREN | tsc | Concat | 6s 34ms/step | 23 | 18.1 | 4.5M | 4438376 | 0.90 | 74.9 (1) |
For multimodal analysis, two CMU datasets were considered:
CMU-MOSEI: 1632 samples were used for training, while the remaining 187 and 465 samples were allocated for validation and testing, respectively trained according to the predetermined protocol.
CMU-MOSI dataset: 1298 samples were categorized with a distribution of 39% positive, 35% negative, and 26% neutral.
We report the 2-class accuracy obtained through 5-fold cross-validation, following the baseline established by A. Zadeh, R. Zellers, E. Pincus, and L. Morency in their work: “MOSI: Multimodal Corpus of Sentiment Intensity and Subjectivity Analysis in Online Opinion Videos,” CoRR, vol. abs/1606.06259, 2016.
Table: Results CMU-MOSI and CMU MOSEI datasets
| Model / Dataset | Mulmix | EmbraceNet | ViT | Meta-Transformer | Minape | SIREN |
|---|---|---|---|---|---|---|
| CMU MOSI | 64.23% | 72.57% | 76.42% | 77.89% | 79.34% | 83.67% |
| CMU MOSEI | 61.48% | 65.32% | 74.15% | 76.04% | 77.21% | 80.61% |
| Average rank | 6 | 5 | 4 | 3 | 2 | 1 |
Note: Table reflects the average accuracy percentage of proposed framework compared to other methods on the CMU MOSI and CMU MOSEI multimodal benchmarks. All experiments performed on a single Nvidia GTX1080Ti 12GB GPU, results calculated over five trials.
To copy and add as the answear The ablation results are reported for Pentostreda dataset. The base results presented in the first row. We have tested the influence of removing the intemediate activtion after the point-wise convolution [3]. We have not notices improvement, in the results. In our work we are using the improved version of activation fuction GeLU. We have conducted the ablation studies by replacing the depth-point block with fully conected bock what resulted in the decrease of the accuracies to 71.3%.
The ablation study results for the Pentostreda dataset are reported below. The baseline results are presented in the first row. We tested the impact of removing the intermediate activation function following the point-wise convolution [3]. No improvement was observed in the results. It's worth noting that in our experiments, we employed an enhanced version of the activation function, GeLU. Additionally, our ablation studies included replacing the depth-point block with a fully connected block, which led to a decrease in accuracy to 71.3%.
Table: Ablation studies on intermediate activation function
| Dataset | Attention Mechanism | Cosine-Decay | Depth-Point | Intermediate Activation | Accuracy |
|---|---|---|---|---|---|
| Pentostreda | × | × | ✓ | ✓ | 74.9% |
| Pentostreda | × | × | ✓ | × | 74.7% |
| Pentostreda | × | × | × | ✓ | 71.3% |
| Pentostreda | × | × | × | × | 71.1% |
We reduced the number of TRANSFORMER_LAYERS from 8 to 2. The outcomes of this modification are detailed in the table below.
Table: Results of the ViT with the reduced size tested on Pentostreda dataset.
| Model | Modality | Fusion-Type | ms/img | Size(MB) | #Params | AUC | Accuracy |
|---|---|---|---|---|---|---|---|
| ViT | t | - | 23 | 104.35 | 9.83M | 0.82 | 38.87% |
| ViT | s | - | 28 | 104.55 | 9.83M | 0.82 | 38.32% |
| ViT | c | - | 22 | 104.35 | 9.86M | 0.84 | 45.62% |
| ViT | ts | Concat | 44 | 209.03 | 19.68M | 0.83 | 40.73% |
| ViT | tc | Concat | 37 | 208.85 | 19.66M | 0.84 | 45.68% |
| ViT | sc | Concat | 39 | 209.03 | 19.68M | 0.84 | 46.51% |
| ViT | tsc | Concat | 64 | 313.02 | 29.43M | 0.84 | 47.34% |
We reduced the number of blocks in the MLP-Mixer from 4 to 1.
Table: Results of the MLP-Mixer with the reduced size tested on Pentostreda dataset.
| Model | Modality | Fusion-Type | ms/img | Size(MB) | #Params | AUC | Accuracy |
|---|---|---|---|---|---|---|---|
| MLP-Mixer | t | - | 10.4 | 7.453 | 0.682M | 0.84 | 42.83% |
| MLP-Mixer | s | - | 14.4 | 8.787 | 0.806M | 0.84 | 45.97% |
| MLP-Mixer | c | - | 10.0 | 7.772 | 0.713M | 0.83 | 41.55% |
| MLP-Mixer | ts | Concat | 17.2 | 13.195 | 1.209M | 0.85 | 49.28% |
| MLP-Mixer | tc | Concat | 18.4 | 11.861 | 1.085M | 0.84 | 43.20% |
| MLP-Mixer | sc | Concat | 17.6 | 13.195 | 1.209M | 0.85 | 50.40% |
| MLP-Mixer | tsc | Concat | 21.2 | 17.255 | 1.581M | 0.86 | 52.13% |
The HA4M dataset focuses on Multi-Modal Monitoring of an assembly task for Human Action Recognition in Manufacturing and includes six modalities: RGB images (r), Depth maps (d), IR images (i), RGB-to-Depth Alignments (g), point maps (p), and Skeleton data (s).
Table: Ablation studies. Influence of the number of modalities tested on HA4M dataset.
| Model | Modality | Fusion-Type | ms/img | Size(MB) | #Params | AUC | Accuracy |
|---|---|---|---|---|---|---|---|
| SIREN | r | - | 9 | 5.5 | 1.4M | 0.85 | 47.3% |
| SIREN | d | - | 10 | 6.3 | 1.5M | 0.85 | 45.3% |
| SIREN | i | - | 11 | 7.1 | 1.6M | 0.84 | 43.3% |
| SIREN | g | - | 12 | 7.9 | 1.7M | 0.85 | 48.4% |
| SIREN | p | - | 13 | 8.3 | 1.8M | 0.84 | 44.4% |
| SIREN | s | - | 16 | 8.3 | 1.9M | 0.84 | 43.0% |
| SIREN | rd | Concat | 19 | 15.1 | 2.8M | 0.87 | 60.4% |
| SIREN | rdi | Concat | 25 | 21.5 | 4.5M | 0.89 | 68.5% |
| SIREN | rdig | Concat | 34 | 27.8 | 6.7M | 0.90 | 73.5% |
| SIREN | rdigp | Concat | 39 | 35.7 | 8.5M | 0.90 | 75.2% |
| SIREN | rdigps | Concat | 48 | 41.3 | 9.7M | 0.90 | 79.2% |
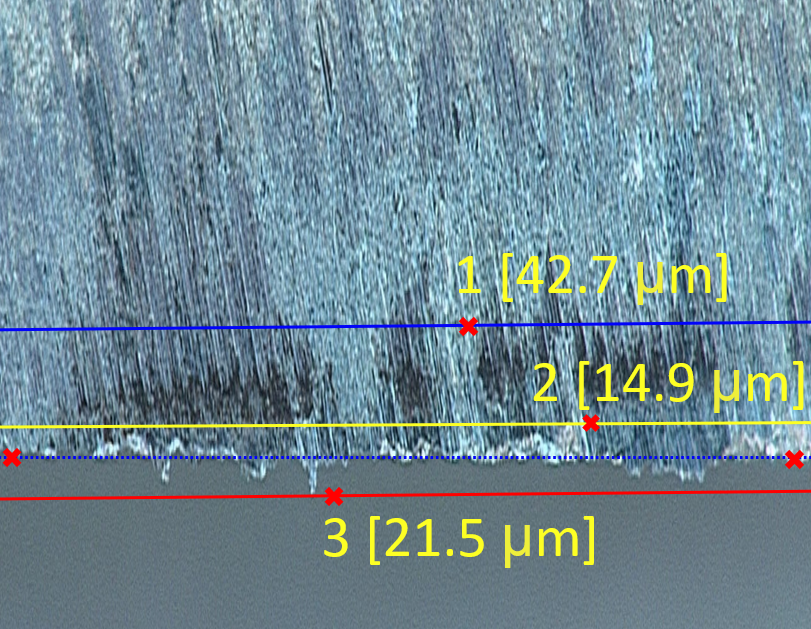
Figure: Visualisation of the base (blue-dotted), tool wear (blue), overhang (red), gaps (yellow) labels used for the regression problem.
Table: Descriptive statistics of the Pentostreda dataaset regression labels.
| Gaps | Flank | Overhang | |
|---|---|---|---|
| Mean | 8.20 | 87.86 | 20.95 |
| Standard Error | 0.67 | 2.01 | 0.52 |
| Median | 5.77 | 82.18 | 18.25 |
| Mode | 0 | 132.72 | 17.72 |
| Standard Deviation | 15.05 | 45.52 | 11.78 |
| Sample Variance | 226.61 | 2072.50 | 138.68 |
| Kurtosis | 48.17 | 4.88 | 2.69 |
| Skewness | 6.08 | 1.45 | 1.40 |
| Range | 154.56 | 324.11 | 76.75 |
| Minimum | 0 | 22.55 | 0 |
| Maximum | 154.56 | 346.66 | 76.75 |
| Sum | 4198.32 | 44985.20 | 10728.76 |
| Count | 512 | 512 | 512 |
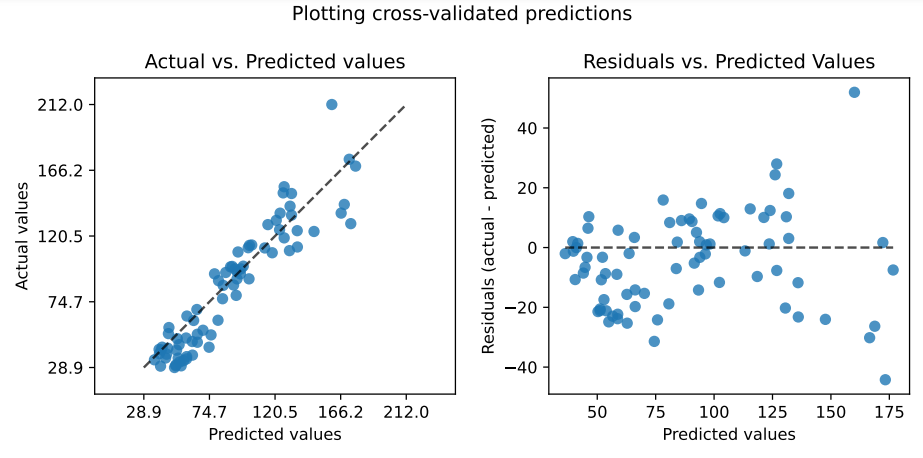
Figure: Visualisation of the actual and the residual flank wear values predicted by Siren on the Pentostreda dataset.
For more visualisation, see: interpretability/ped
The total weights of the patch embedding layers in a Siren with a patch of 16 are visualized. While these layers essentially act as crude edge detectors, the industrial nature of the Mudestreda dataset prevents any discernible patterns from emerging. Interestingly, a number of filters bear a striking resemblance to noise, indicating the potential requirement for increased regularization.
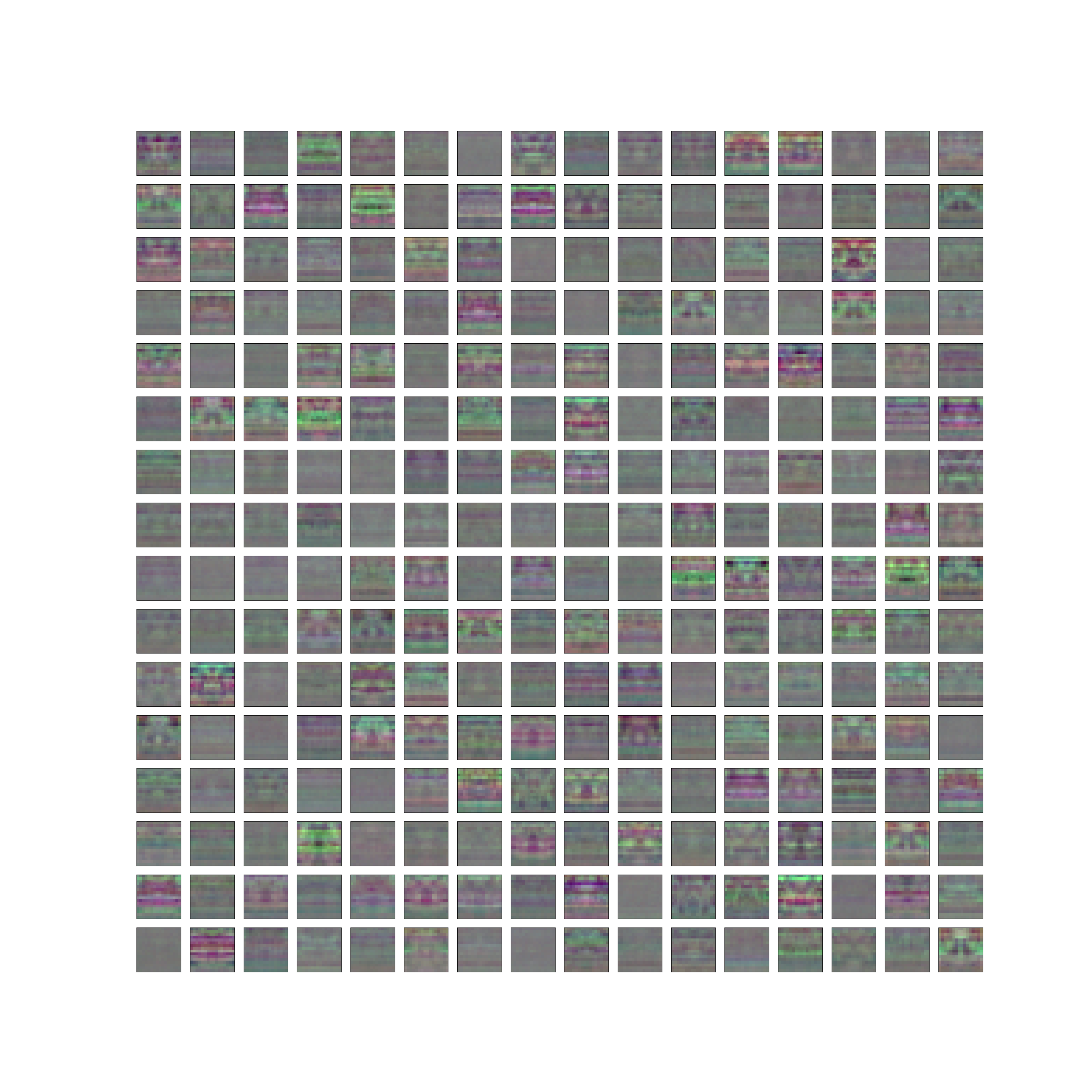
Figure: The total weights of the patch embedding layers in a SIREN with a patch of 8 for spectrogram F_x.

Figure: The total weights of the patch embedding layers in a SIREN with a patch of 8 for spectrogram F_y.

Figure: The total weights of the patch embedding layers in a SIREN with a patch of 8 for spectrogram F_z.

Figure: The total weights of the patch embedding layers in a SIREN with a patch of 8 for chip images.

Figure: The total weights of the patch embedding layers in a SIREN with a patch of 8 for tool images.
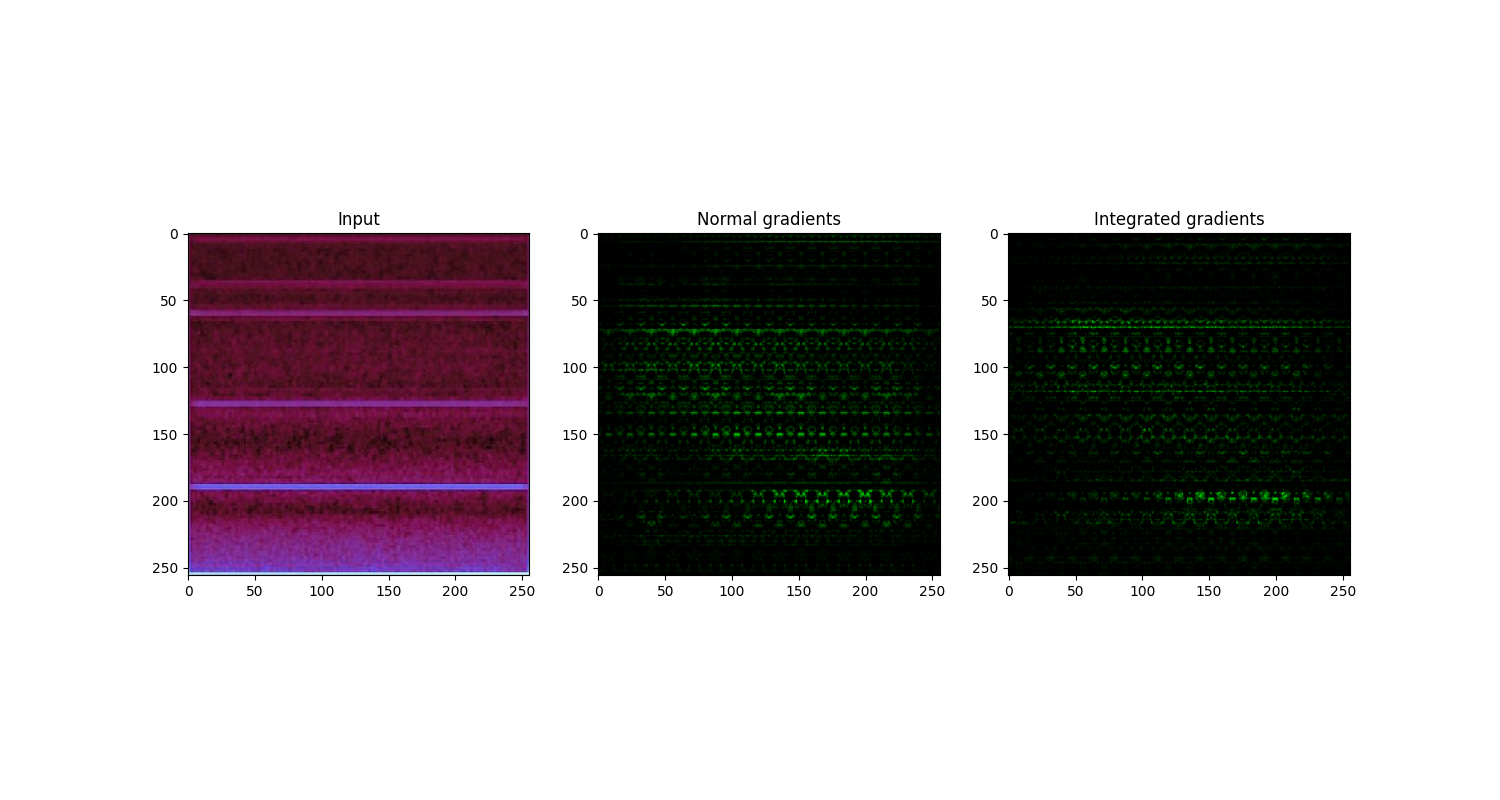
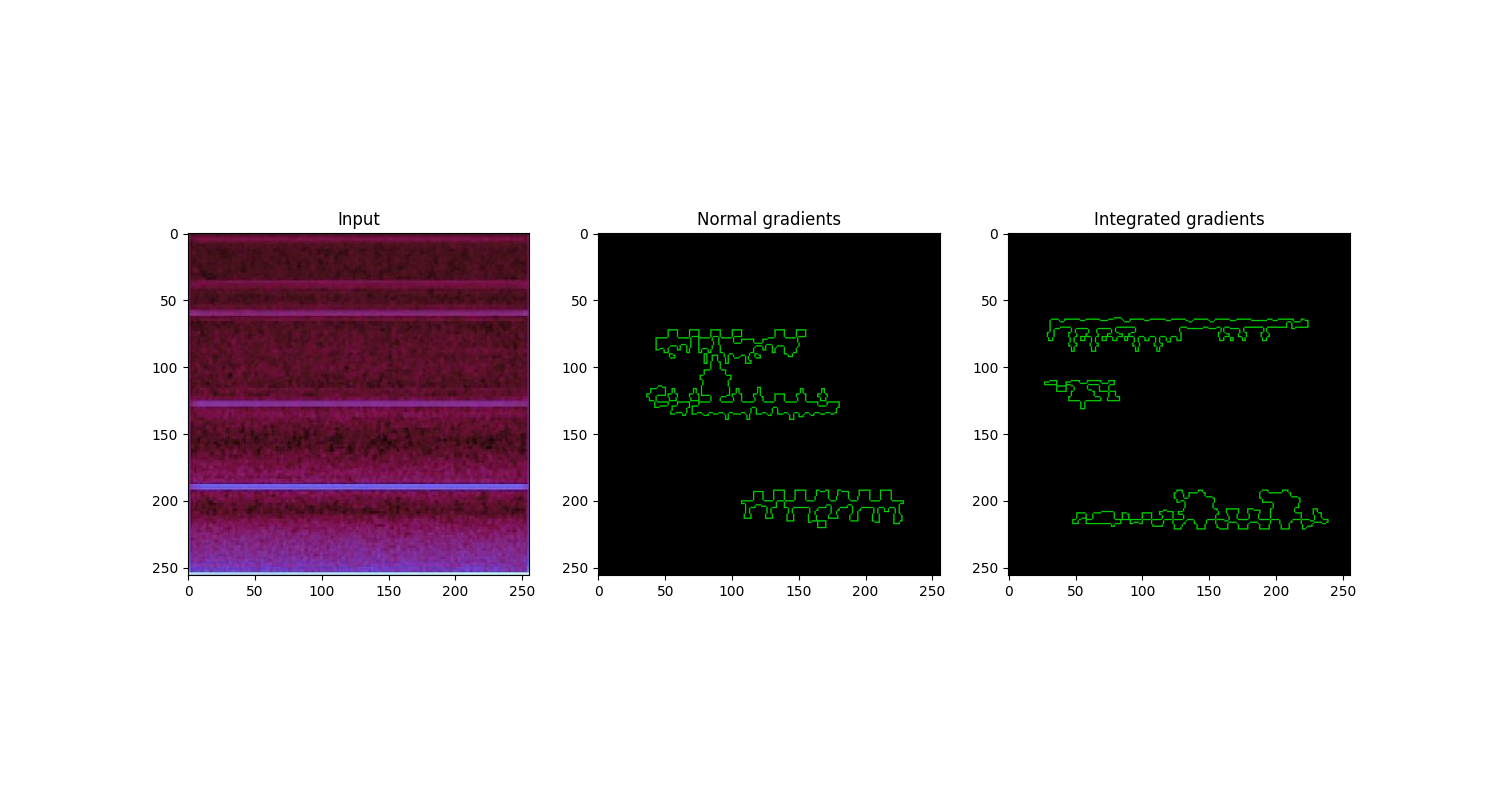
Figure: Visual comparison of normal gradient and integrated gradient on a used tool blade image. The normal gradient and integrated gradient images offer pixel-wise and area-wise importance visualization, respectively.
For more visualisation, see: interpretability
This section provides instructions for contributing to the project, including how to report bugs, submit feature requests, or contribute code. It should also include information about the development process and any guidelines for contributing.
Pull requests are great. For major changes, please open an issue first to discuss what you would like to change.
Please make sure to update tests as appropriate.