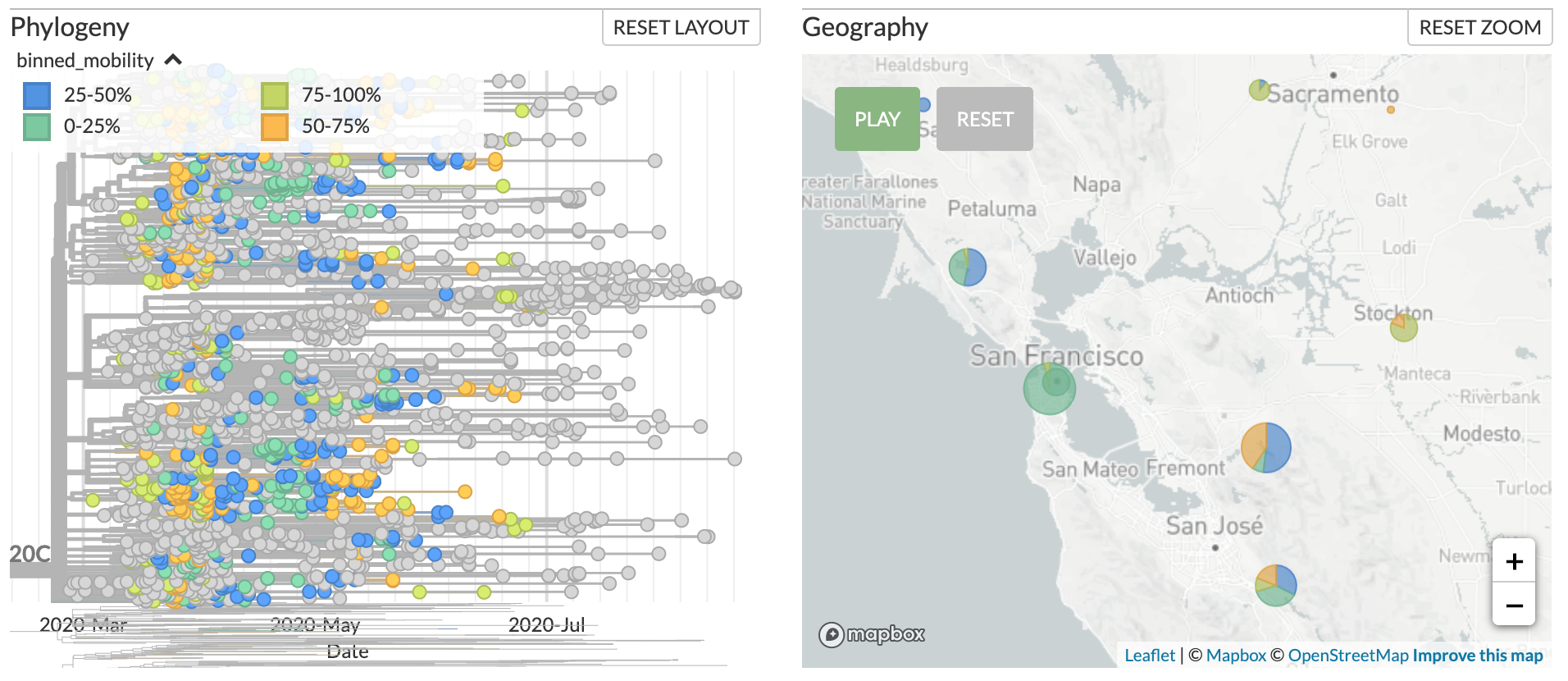
This repo provides a minimal, proof-of-concept analysis for how one might annotate phylogenies inferred from SARS-CoV-2 genomes with mobility data to better understand how changes in mobility are related to viral transmission.
View the precompiled notebook, to get a sense of how the analysis works.
First, install miniconda. Then, create the associated conda environment and activate it.
conda env create -f=environment.yaml
conda activate mobilityStart a Jupyter lab session and run the entire notebook from scratch.
jupyter lab 2020-09-03-annotate-sars-cov-2-phylogeny-with-mobility-data.ipynbDownload the precalculated data file to drag-and-drop onto the corresponding phylogeny of Californian SARS-CoV-2 genomes. An example of the resulting annotation is included below.