This repository provides an overview of all components from the paper Scaling Data-Constrained Language Models. Talks on the paper:
- 65min talk by Niklas Muennighoff
- 60min talk by Niklas Muennighoff
- 63min talk by Alexander M. Rush
- 25min talk by Alexander M. Rush
- 60min slide deck used by Niklas Muennighoff
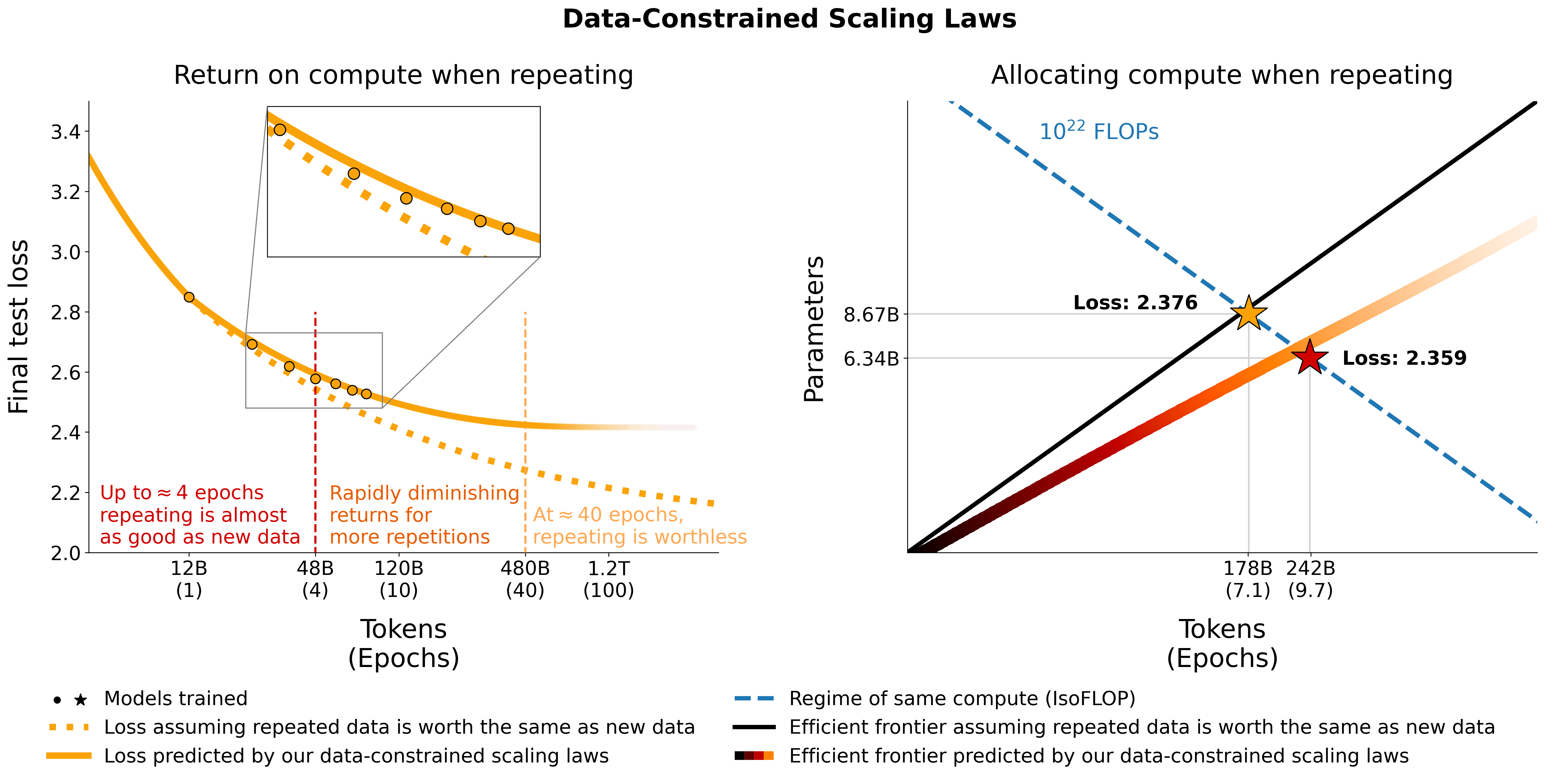
We investigate scaling language models in data-constrained regimes. We run a large set of experiments varying the extent of data repetition and compute budget, ranging up to 900 billion training tokens and 9 billion parameter models. Based on our runs we propose and empirically validate a scaling law for compute optimality that accounts for the decreasing value of repeated tokens and excess parameters. We also experiment with approaches mitigating data scarcity, including augmenting the training dataset with code data, perplexity-filtering and deduplication. Models and datasets from our 400 training runs are available via this repository.
We experiment with repeating data on C4 and the non-deduplicated English split of OSCAR. For each dataset, we download the data and turn it into a single jsonl file, c4.jsonl and oscar_en.jsonl respectively.
Then we decide on the amount of unique tokens and the respective number of samples we need from the dataset. Note that C4 has 478.625834583 tokens per sample and OSCAR has 1312.0951072 with the GPT2Tokenizer. This was calculated by tokenizing the entire dataset and dividing the number of tokens by the number of samples. We use these numbers to calculate the needed samples.
For example, for 1.9B unique tokens, we need 1.9B / 478.625834583 = 3969697.96178 samples for C4 and 1.9B / 1312.0951072 = 1448065.76107 samples for OSCAR. To tokenize the data, we first need to clone the Megatron-DeepSpeed repository and follow its setup guide. We then select these samples and tokenize them as follows:
C4:
head -n 3969698 c4.jsonl > c4_1b9.jsonl
python Megatron-DeepSpeed/tools/preprocess_data_many_cores.py \
--input c4_1b9.jsonl \
--output-prefix gpt2tok_c4_en_1B9 \
--dataset-impl mmap \
--tokenizer-type PretrainedFromHF \
--tokenizer-name-or-path gpt2 \
--append-eod \
--workers 64OSCAR:
head -n 1448066 oscar_en.jsonl > oscar_1b9.jsonl
python Megatron-DeepSpeed/tools/preprocess_data_many_cores.py \
--input oscar_1b9.jsonl \
--output-prefix gpt2tok_oscar_en_1B9 \
--dataset-impl mmap \
--tokenizer-type PretrainedFromHF \
--tokenizer-name-or-path gpt2 \
--append-eod \
--workers 64where gpt2 points to a folder containing all files from https://huggingface.co/gpt2/tree/main. By using head we make sure that different subsets will have overlapping samples to reduce randomness.
For evaluation during training and the final evaluation, we use the validation set for C4:
from datasets import load_dataset
load_dataset("c4", "en", split="validation").to_json("c4-en-validation.json")python Megatron-DeepSpeed/tools/preprocess_data_many_cores.py \
--input c4-en-validation.jsonl \
--output-prefix gpt2tok_c4validation_rerun \
--dataset-impl mmap \
--tokenizer-type PretrainedFromHF \
--tokenizer-name-or-path gpt2 \
--append-eod \
--workers 2For OSCAR which has no official validation set we take a part of the training set by doing tail -364608 oscar_en.jsonl > oscarvalidation.jsonl and then tokenize it as follows:
python Megatron-DeepSpeed/tools/preprocess_data_many_cores.py --input oscarvalidation.jsonl --output-prefix gpt2tok_oscarvalidation --dataset-impl mmap --tokenizer-type PretrainedFromHF --tokenizer-name-or-path gpt2 --append-eod --workers 2We have uploaded several preprocessed subsets for usage with megatron:
- C4: https://huggingface.co/datasets/datablations/c4-subsets
- OSCAR: https://huggingface.co/datasets/datablations/oscar-subsets
Some bin files were too large for git and thus split using e.g. split --number=l/40 gpt2tok_c4_en_1B9.bin gpt2tok_c4_en_1B9.bin. and split --number=l/40 gpt2tok_oscar_en_1B9.bin gpt2tok_oscar_en_1B9.bin.. To use them for training you need to cat them together again using cat gpt2tok_c4_en_1B9.bin.* > gpt2tok_c4_en_1B9.bin and cat gpt2tok_oscar_en_1B9.bin.* > gpt2tok_oscar_en_1B9.bin.
We experiment with mixing code with the natural language data using the Python split from the the-stack-dedup. We download the data, turn it into a single jsonl file and preprocess it using the same approach as outlined above.
We have uploaded the preprocessed version for usage with megatron here: https://huggingface.co/datasets/datablations/python-megatron. We have split the bin file using split --number=l/40 gpt2tok_python_content_document.bin gpt2tok_python_content_document.bin., so you need to cat them together again using cat gpt2tok_python_content_document.bin.* > gpt2tok_python_content_document.bin for training.
We create versions of C4 and OSCAR with perplexity and deduplication-related filtering metadata:
- C4: https://huggingface.co/datasets/datablations/c4-filter
- OSCAR: https://huggingface.co/datasets/datablations/oscar-filter
To recreate these metadata datasets there are instructions at filtering/README.md.
We provide the tokenized versions that can be used for training with Megatron at:
- C4: https://huggingface.co/datasets/datablations/c4-filter-megatron
- OSCAR: https://huggingface.co/datasets/datablations/oscar-filter-megatron
.bin files were split using something like split --number=l/10 gpt2tok_oscar_en_perplexity_25_text_document.bin gpt2tok_oscar_en_perplexity_25_text_document.bin., so you need to concatenate them back together via cat gpt2tok_oscar_en_perplexity_25_text_document.bin. > gpt2tok_oscar_en_perplexity_25_text_document.bin.
To recreate the tokenized versions given the metadata dataset,
- OSCAR:
- Deduplication: See
filtering/deduplication/filter_oscar_jsonl.py - Perplexity: See below.
- Deduplication: See
- C4:
- Deduplication: See below.
- Perplexity: See below.
To create the perplexity percentiles, follow the below instructions.
C4:
from datasets import load_dataset
import numpy as np
ds = load_dataset("datablations/c4-filter", streaming=False, num_proc=128)
p_25 = np.percentile(ds["train"]["perplexity"], 25)
p_50 = np.percentile(ds["train"]["perplexity"], 50)
p_75 = np.percentile(ds["train"]["perplexity"], 75)
# 25 - 75th percentile
ds["train"].filter(lambda x: p_25 < x["perplexity"] < p_75, num_proc=128).to_json("c4_perplexty2575.jsonl", num_proc=128, force_ascii=False)
# 25th percentile
ds["train"].filter(lambda x: x["perplexity"] < p_25, num_proc=128).to_json("c4_perplexty25.jsonl", num_proc=128, force_ascii=False)
# 50th percentile
ds["train"].filter(lambda x: x["perplexity"] < p_50, num_proc=128).to_json("c4_perplexty50.jsonl", num_proc=128, force_ascii=False)OSCAR:
from datasets import load_dataset
import numpy as np
ds = load_dataset("datablations/oscar-filter", use_auth_token=True, streaming=False, num_proc=128)
p_25 = np.percentile(ds["train"]["perplexity_score"], 25)
p_50 = np.percentile(ds["train"]["perplexity_score"], 50)
# 25th percentile
ds["train"].filter(lambda x: x["perplexity_score"] < p_25, num_proc=128).remove_columns(['meta', 'perplexity_score', 'text_length', 'url', 'domain', 'dup_ratio', 'pairs', 'repetitions', 'included_in_dedup', 'cluster', 'id']).to_json("oscar_perplexity25.jsonl", num_proc=128, force_ascii=False)
# 50th percentile
ds["train"].filter(lambda x: x["perplexity_score"] < p_50, num_proc=128).remove_columns(['meta', 'perplexity_score', 'text_length', 'url', 'domain', 'dup_ratio', 'pairs', 'repetitions', 'included_in_dedup', 'cluster', 'id']).to_json("oscar_perplexity50.jsonl", num_proc=128, force_ascii=False)You can then tokenize the resulting jsonl files for training with Megatron as described in the Repeating section.
C4:
For C4 you just need to remove all samples where the repetitions field is populated, via e.g.
from datasets import load_dataset
import numpy as np
ds = load_dataset("datablations/c4-dedup", use_auth_token=True, streaming=False, num_proc=128)
ds.filter(lambda x: not(x["repetitions"]).to_json('c4_dedup.jsonl', num_proc=128, force_ascii=False)OSCAR:
For OSCAR we provide a script at filtering/filter_oscar_jsonl.py to create the deduplicated dataset given the dataset with filtering metadata.
You can then tokenize the resulting jsonl files for training with Megatron as described in the Repeating section.
All models can be downloaded at https://huggingface.co/datablations.
Models are generally named as follows: lm1-{parameters}-{tokens}-{unique_tokens}, specifically individual models in the folders are named as: {parameters}{tokens}{unique_tokens}{optional specifier}, for example 1b12b8100m would be 1.1 billion params, 2.8 billion tokens, 100 million unique tokens. The xby (1b1, 2b8 etc.) convention introduces some ambiguity whether numbers belong to parameters or tokens, but you can always check the sbatch script in the respective folder to see the exact parameters / tokens / unique tokens. If you want to convert models that have not yet been converted to huggingface/transformers, you can follow the instructions in Training.
The easiest way to download a single model is e.g.:
GIT_LFS_SKIP_SMUDGE=1 git clone https://huggingface.co/datablations/lm1-misc
cd lm1-misc; git lfs pull --include 146m14b400m/global_step21553If this takes too long, you can also use wget to directly download individual files from the folder, e.g.:
wget https://huggingface.co/datablations/lm1-misc/resolve/main/146m14b400m/global_step21553/bf16_zero_pp_rank_0_mp_rank_00_optim_states.ptFor models corresponding to the experiments in the paper, consult the following repositories:
- IsoLoss experiments & double descent data from the Appendix:
- https://huggingface.co/datablations/lm1-misc
lm1-misc/*dedup*for deduplication comparison on 100M unique tokens in the appendix
- IsoFLOP experiments:
- C4:
- https://huggingface.co/datablations/lm1-8b7-178b-c4-repetitions
- https://huggingface.co/datablations/lm1-4b2-84b-c4-repetitions
- https://huggingface.co/datablations/lm1-4b2-84b-c4seeds
- https://huggingface.co/datablations/lm1-2b8-55b-c4-repetitions
- https://huggingface.co/datablations/lm1-2b8-55b-c4seeds
- OSCAR:
- https://huggingface.co/datablations/lm1-8b7-178b-oscar-repetitions
- https://huggingface.co/datablations/lm1-4b2-84b-oscar-repetitions
- https://huggingface.co/datablations/lm1-4b2-84b-oscarseeds
- https://huggingface.co/datablations/lm1-2b8-55b-oscar-repetitions
- https://huggingface.co/datablations/lm1-2b8-55b-oscarseeds
- C4:
- Code augmentation:
- Perplexity-filtering:
- C4:
- https://huggingface.co/datablations/lm1-4b2-84b-c4-perplexity
- https://huggingface.co/datablations/lm1-2b8-55b-c4-perplexity
- https://huggingface.co/datablations/lm1-2b8-84b-c4-perplexity
- These are not referenced anywhere in the paper, but could be used
- OSCAR:
- C4:
- Deduplication:
- ROOTS filtering
- muP (Appendix):
Other models not analysed in the paper:
- UL2: https://huggingface.co/datablations/lm5-2b8-55b-c4
- All checkpoints of the c4 8b7 model trained for 1 epoch: https://huggingface.co/datablations/lm1-8b7-176b-c4-ckpts
- Adam beta2 ablation: https://huggingface.co/datablations/lm1-8b7-12b-beta
- Warmup ablation: https://huggingface.co/datablations/lm1-83m-20b-nowarmup
- An experiment pre-training only on instruction datasets: https://huggingface.co/datablations/lm1-2b8-55b-realtasky
- Misc runs on The Pile: https://huggingface.co/datablations/lm1-misc-pile
- Misc runs on OSCAR: https://huggingface.co/datablations/lm1-misc-oscar
- IsoFLOP for 1.1B on C4: https://huggingface.co/datablations/lm1-1b1-21b-c4 & https://huggingface.co/datablations/lm1-1b1-21b-c4-repetitions & https://huggingface.co/datablations/lm1-1b1-21b-c4seeds
We train models with our fork of Megatron-DeepSpeed that works with AMD GPUs (via ROCm): https://github.com/TurkuNLP/Megatron-DeepSpeed If you would like to use NVIDIA GPUs (via cuda), you can use the original library: https://github.com/bigscience-workshop/Megatron-DeepSpeed
You need to follow the setup instructions of either repository to create your environment (Our setup specific to LUMI is detailed in training/megdssetup.md).
Each model folder contains an sbatch script that was used to train the model. You can use these as a reference to train your own models adapting the necessary environment variables. The sbatch scripts reference some additional files:
*txtfiles that specify the data paths. You can find them atutils/datapaths/*, however, you will likely need to adapt the path to point to your dataset.model_params.sh, which is atutils/model_params.shand contains architecture presets.launch.shthat you can find attraining/launch.sh. It contains commands specific to our setup, which you may want to remove.
After training you can convert your model to transformers with e.g. python Megatron-DeepSpeed/tools/convert_checkpoint/deepspeed_to_transformers.py --input_folder global_step52452 --output_folder transformers --target_tp 1 --target_pp 1.
For repeat models, we also upload their tensorboards after training using e.g. tensorboard dev upload --logdir tensorboard_8b7178b88boscar --name "tensorboard_8b7178b88boscar", which makes them easy to use for visualization in the paper.
For the muP ablation in the Appendix we use the script at training_scripts/mup.py. It contains setup instructions.
You can use our formula to compute the expected loss given parameters, data and unique tokens as follows:
import numpy as np
func = r"$L(N,D,R_N,R_D)=E + \frac{A}{(U_N + U_N * R_N^* * (1 - e^{(-1*R_N/(R_N^*))}))^\alpha} + \frac{B}{(U_D + U_D * R_D^* * (1 - e^{(-1*R_D/(R_D^*))}))^\beta}$"
a, b, e, alpha, beta, rd_star, rn_star = [6.255414, 7.3049974, 0.6254804, 0.3526596, 0.3526596, 15.387756, 5.309743]
A = np.exp(a)
B = np.exp(b)
E = np.exp(e)
G = ((alpha*A)/(beta*B))**(1/(alpha+beta))
def D_to_N(D):
return (D * G)**(beta/alpha) * G
def scaling_law(N, D, U):
"""
N: number of parameters
D: number of total training tokens
U: number of unique training tokens
"""
assert U <= D, "Cannot have more unique tokens than total tokens"
RD = np.maximum((D / U) - 1, 0)
UN = np.minimum(N, D_to_N(U))
RN = np.maximum((N / UN ) - 1, 0)
L = E + A/(UN + UN*rn_star*(1-np.exp(-1*RN/rn_star)))**alpha + B / (U + U * rd_star * (1 - np.exp(-1*RD/(rd_star))))**beta
return L
# Models in Figure 1 (right):
print(scaling_law(6.34e9, 242e9, 25e9)) # 2.2256440889984477 # <- This one is better
print(scaling_law(8.67e9, 178e9, 25e9)) # 2.2269634075087867Note that the actual loss value is unlikely to be useful, but rather the trend of the loss as e.g. the number of parameters increases or to compare two models like in the example above. To compute the optimal allocation, you can use a simple grid search:
def chinchilla_optimal_N(C):
a = (beta)/(alpha+beta)
N_opt = G*(C/6)**a
return N_opt
def chinchilla_optimal_D(C):
b = (alpha)/(alpha+beta)
D_opt = (1/G)*(C/6)**b
return D_opt
def optimal_allocation(C, U_BASE):
"""Compute optimal number of parameters and tokens to train for given a compute & unique data budget"""
N_BASE = chinchilla_optimal_N(C)
D_BASE = chinchilla_optimal_D(C)
min_l = float("inf")
for i in np.linspace(1.0001, 3, 500):
D = D_BASE*i
U = min(U_BASE, D)
N = N_BASE/i
new_l = scaling_law(N, D, U)
if new_l < min_l:
min_l, min_t, min_s = new_l, D, N
D = D_BASE/i
U = min(U_BASE, D)
N = N_BASE*i
new_l = scaling_law(N, D, U)
if new_l < min_l:
min_l, min_t, min_s = new_l, D, N
return min_l, min_t, min_s
_, min_t, min_s = optimal_allocation(10**22, 25e9)
print(f"Optimal configuration: {min_t} tokens, {min_t/25e9} epochs, {min_s} parameters")
# -> 237336955477.55075 tokens, 9.49347821910203 epochs, 7022364735.879969 parameters
# We went more extreme in Figure 1 to really put our prediction of "many epochs, fewer params" to the testIf you derive a closed-form expression for the optimal allocation instead of the above grid search, please let us know :) We fit data-constrained scaling laws & the C4 scaling coefficients using the code at utils/parametric_fit.ipynb equivalent to this colab.
- Follow the instructions in the
Training>Regular modelssection to setup a training environment. - Install the evaluation harness:
pip install git+https://github.com/EleutherAI/lm-evaluation-harness.git. We used version 0.2.0, but newer versions should work as well. - Run
sbatch utils/eval_rank.shmodifying the necessary variables in the script first - After running, we convert each file to csv using
python Megatron-DeepSpeed/tasks/eval_harness/report-to-csv.py outfile.json
- Clone the
addtasksbranch of the evaluation harness:git clone -b addtasks https://github.com/Muennighoff/lm-evaluation-harness.git - Setup an environment with
cd lm-evaluation-harness; pip install -e ".[dev]"; pip uninstall -y promptsource; pip install git+https://github.com/Muennighoff/promptsource.git@tr13i.e. all requirements except promptsource, which is installed from a fork with the correct prompts - Make sure your checkpoint path is a transformers checkpoint path
- Run
sbatch utils/eval_generative.shmodifying the necessary variables in the script first - After running, we merge the generation files using
python utils/merge_generative.pyand then convert them to csv withpython utils/csv_generative.py merged.json
- Clone the
babibranch of the evaluation harness:git clone -b babi https://github.com/Muennighoff/lm-evaluation-harness.git(Note that this branch is not compatible with theaddtasksbranch for generative tasks as it stems fromEleutherAI/lm-evaluation-harness, whileaddtasksis based onbigscience/lm-evaluation-harness) - Setup an environment with
cd lm-evaluation-harness; pip install -e ".[dev]" - Make sure your checkpoint path is a transformers checkpoint with tokenizer files (If you trained a gpt2 model like all models in this work, it's just the files from here: https://huggingface.co/gpt2)
- Run
sbatch utils/eval_babi.shmodifying the necessary variables in the script first
- Figure 1:
plotstables/return_alloc.pdf,plotstables/return_alloc.ipynb, colab - Figure 2:
plotstables/dataset_setup.pdf,plotstables/dataset_setup.ipynb, colab - Figure 3:
plotstables/contours.pdf,plotstables/contours.ipynb, colab - Figure 4:
plotstables/isoflops_training.pdf,plotstables/isoflops_training.ipynb, colab - Figure 5:
plotstables/return.pdf,plotstables/return.ipynb, colab - Figure 6 (Left):
plotstables/strategies.pdf,plotstables/strategies.drawio - Figure 6 (Right):
plotstables/beyond.pdf,plotstables/beyond.ipynb, colab - Figure 7:
plotstables/cartoon.pdf,plotstables/cartoon.pptx - Figure 8:
plotstables/isoloss_400m1b5.pdf& same colab as Figure 3 - Figure 9 - 11:
plotstables/mup.pdf,plotstables/dd.pdf,plotstables/dedup.pdf,plotstables/mup_dd_dd.ipynb, colab - Figure 12:
plotstables/isoloss_alphabeta_100m.pdf& same colab as Figure 3 - Figure 13:
plotstables/galactica.pdf,plotstables/galactica.ipynb, colab - Figure 14 - 17:
training_c4.pdf,validation_c4oscar.pdf,training_oscar.pdf,validation_epochs_c4oscar.pdf& same colab as Figure 4 - Figure 18:
plotstables/perplexity_histogram.pdf,plotstables/perplexity_histogram.ipynb - Figure 19 - 20:
plotstabls/validation_c4py.pdf,plotstables/training_validation_filter.pdf,plotstables/beyond_losses.ipynb& colab - Figure 21 - 39: Manual
- Table 1 - 3: Manual; The experiments for Table 1 are in
utils/parametric_fit.ipynbequivalent to this colab. - Table 4 - 9:
plotstables/repetition.ipynb& colab - Table 10 - 11:
plotstables/python.ipynb& colab - Table 12: Manual
- Table 13 - 14:
plotstables/filtering.ipynb& colab - Table 15: Manual
All models & code are licensed under Apache 2.0. Filtered datasets are released with the same license as the datasets they stem from.
@article{muennighoff2023scaling,
title={Scaling Data-Constrained Language Models},
author={Muennighoff, Niklas and Rush, Alexander M and Barak, Boaz and Scao, Teven Le and Piktus, Aleksandra and Tazi, Nouamane and Pyysalo, Sampo and Wolf, Thomas and Raffel, Colin},
journal={arXiv preprint arXiv:2305.16264},
year={2023}
}