This is the official code for our MedIA paper:
Nuclei Segmentation with Point Annotations from Pathology Images via Self-Supervised Learning and Co-Training
Yi Lin*, Zhiyong Qu*, Hao Chen, Zhongke Gao, Yuexiang Li, Lili Xia, Kai Ma, Yefeng Zheng, Kwang-Ting Cheng
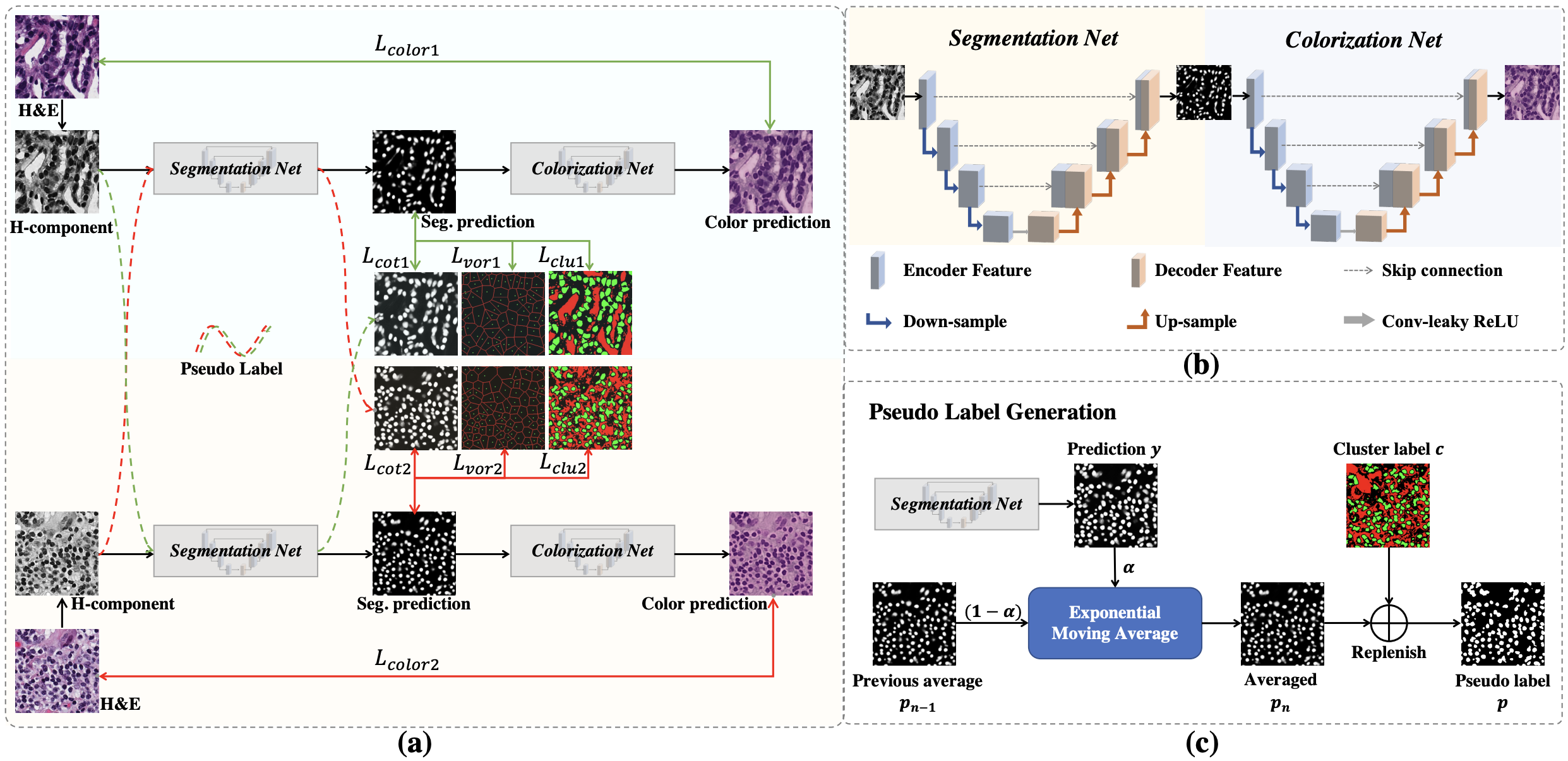
In this work, we propose a weakly-supervised learning method for nuclei segmentation that only requires point annotations for training. The proposed method achieves label propagation in a coarse-to-fine manner as follows. First, coarse pixel-level labels are derived from the point annotations based on the Voronoi diagram and the k-means clustering method to avoid overfitting. Second, a co-training strategy with an exponential moving average method is designed to refine the incomplete supervision of the coarse labels. Third, a self-supervised visual representation learning method is tailored for nuclei segmentation of pathology images that transforms the hematoxylin component images into the H&E stained images to gain better understanding of the relationship between the nuclei and cytoplasm.
Please clone the following repositories:
git clone https://github.com/hust-linyi/SC-Net.git
pip install -r requirements.txt
Please refer to dataloaders/prepare_data.py for the pre-processing of the datasets.
- Configure your own parameters in opinions.py, including the dataset path, the number of GPUs, the number of epochs, the batch size, the learning rate, etc.
- Run the following command to train the model:
python train.py
Run the following command to test the model:
python test.py
Please cite the paper if you use the code.
@article{lin2023nuclei,
title={Nuclei segmentation with point annotations from pathology images via self-supervised learning and co-training},
author={Lin, Yi and Qu, Zhiyong and Chen, Hao and Gao, Zhongke and Li, Yuexiang and Xia, Lili and Ma, Kai and Zheng, Yefeng and Cheng, Kwang-Ting},
journal={Medical Image Analysis},
pages={102933},
year={2023},
publisher={Elsevier}
}