thresholdmodeling: A Python package for modeling excesses over a threshold using the Peak-Over-Threshold Method and the Generalized Pareto Distribution
This package is intended for those who wish to conduct an extreme values analysis. It provides the whole toolkit necessary to create a threshold model in a simple and efficient way, presenting the main methods towards the Peak-Over-Threshold method and the fit in the Generalized Pareto Distribution.
In this repository you can find the main files of the package, the Functions Documenation, the dataset used in some examples, the paper submitted to the Jounal of Open Source Software and some tutorials.
It is necessary to have internet connection and use Anaconda distribution (Python 3).
-
For installing Anaconda on Linux, go to this link. For installing on Windows, go to this one. For istalling on macOS, go to this one.
-
For creating your own environment by using the terminal or Anaconda Prompt, go here.
Firstly, it will necessary to install R on your environment and considering that rpy2 (a python dependency package for thresholdmodeling) does not have Windows support, installing it from pip install thresholdmodeling will result in an error, the same occurs with pip install rpy2. Then, it is necessary to download it from an unuofficial website:
https://www.lfd.uci.edu/~gohlke/pythonlibs/
Here, you must find the rpy2 realese which works on your machine and install it manually going to the download folder with the Anaconda Prompt and run this line, for example (it will depend on the name of the downloaded file):
pip install rpy2‑2.9.5‑cp37‑cp37m‑win_amd64.whl
Or you can install it from the the Anaconda Prompt by activating your environment and running:
conda activate my_env
conda install r
conda install -c r rpy2=2.9.4
After that, rpy2 and R will be installed on your machine. Follow the next steps.
For installing the package just use the following command on your Anaconda Prompt (it is already in PyPi):
pip install thresholdmodeling
The others Python dependencies for runing the software will install automatically with this command.
Once the package is installed, it is necessary to run these lines on your IDE for installing POT R package (package that our software uses by means of rpy2 for computing GPD estimatives):
from rpy2.robjects.packages import importr
import rpy2.robjects.packages as rpackages
base = importr('base')
utils = importr('utils')
utils.chooseCRANmirror(ind=1)
utils.install_packages('POT') #installing POT packageFirstly, run this lines on your terminal in order to install R and rpy2 package on your environment:
conda activate my_env (my_env is your environment name)
conda install r
conda install -c r rpy2=2.9.4
After installing R and rpy2, find your anaconda directory, and find the actual environment folder. It should be somewhere like ~/anaconda3/envs/my_env. Open the terminal in this folder and run this line (the others dependencies will automatically install):
pip install thresholdmodeling
Once the package is installed, it is necessary to run this lines on your IDE for installing POT R package (package that our software uses by means of rpy2 for computing GPD estimatives):
from rpy2.robjects.packages import importr
import rpy2.robjects.packages as rpackages
base = importr('base')
utils = importr('utils')
utils.chooseCRANmirror(ind=1)
utils.install_packages('POT') #installing POT package
Or, it is possible to download this [file](https://github.com/iagolemos1/thresholdmodeling/blob/master/install_pot.py) in order to run it in yout IDE and installing ``POT``.In the file example it is possible to see how the package should be used. In Functions Documenation it may be seen a complete documentation on how to use the functions presented in the package.
In order to present a tutorial on how to use the package and its results, a guide is presented below, using the example on the Coles's book with the Daily Rainfall in South-West England dataset.
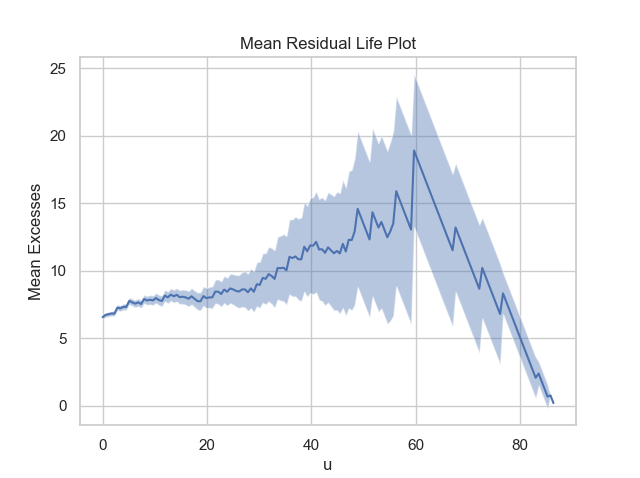
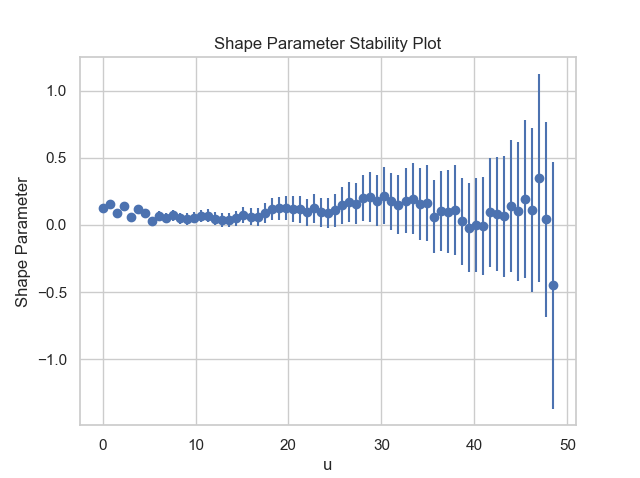
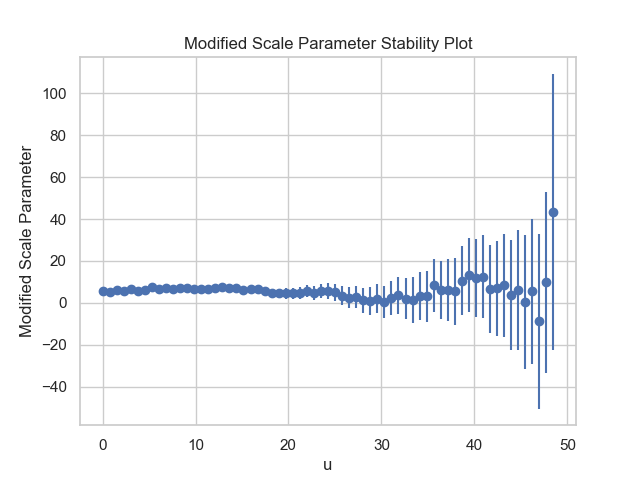
Firstly, it is necessary to conduct a threshold value analysis using the first two functions of the package: MRL and Parameter_Stability_Plot, in order to select a reasonable threshold value.
Runing this:
from thresholdmodeling import thresh_modeling #importing package
import pandas as pd #importing pandas
url = 'https://raw.githubusercontent.com/iagolemos1/thresholdmodeling/master/dataset/rain.csv' #saving url
df = pd.read_csv(url, error_bad_lines=False) #getting data
data = df.values.ravel() #turning data into an array
thresh_modeling.MRL(data, 0.05)
thresh_modeling.Parameter_Stability_plot(data, 0.05)The results must be:
Then, by analysing the three graphics, it is reasonable taking the threshold value as 30.
Once the threshold value is defined, it is possible to fit the dataset to a GPD model by using the function gpdfitrunning the following line and using the maximum likelihood estimation method:
thresh_modeling.gpdfit(data, 30, 'mle')The results must be in Terminal like:
Estimator: MLE
Deviance: 970.1874
AIC: 974.1874
Varying Threshold: FALSE
Threshold Call: 30L
Number Above: 152
Proportion Above: 0.0087
Estimates
scale shape
7.4411 0.1845
Standard Error Type: observed
Standard Errors
scale shape
0.9587 0.1012
Asymptotic Variance Covariance
scale shape
scale 0.91920 -0.06554
shape -0.06554 0.01025
Optimization Information
Convergence: successful
Function Evaluations: 14
Gradient Evaluations: 6
These are the GPD model estimatives using the maximum likelihood estimator.
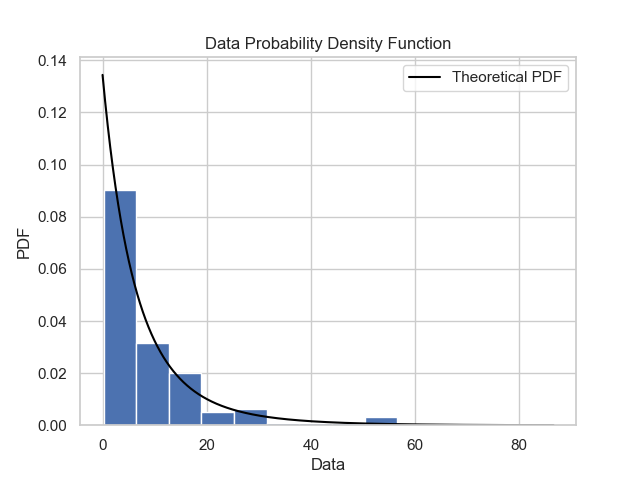
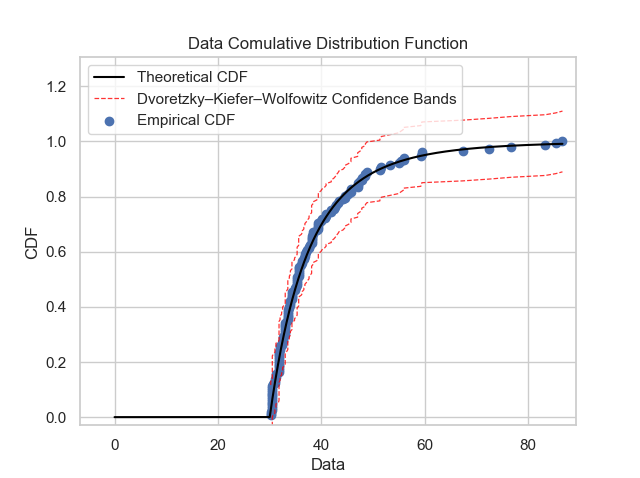
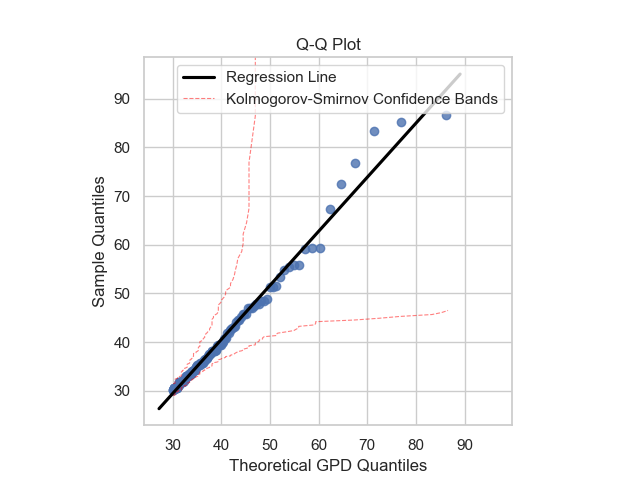
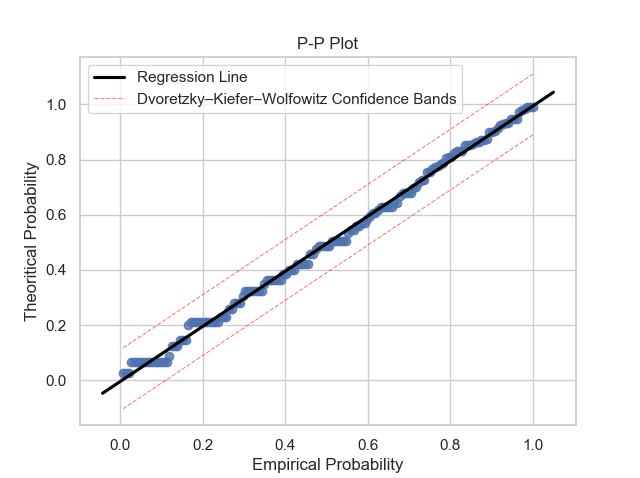
Once the GPD model is defined, it is necessary to verify if the model is reasonable and describes well the empirical observations. Plots like probability density function, cumulative distribution function, quantile-quantile and probability-probability can show to us if the model is good. It is possible to obtain these plots using some functions of the package: gpdpdf, gpdcdf, qqplot and ppplot. By running these lines:
thresh_modeling.gpdpdf(data, 30, 'mle', 'sturges', 0.05)
thresh_modeling.gpdcdf(data, 30, 'mle', 0.05)
thresh_modeling.qqplot(data,30, 'mle', 0.05)
thresh_modeling.ppplot(data, 30, 'mle', 0.05)The results must be:
Once it is possible to verifiy that the theoretical model describes very well the empirical observations, the next step is to use the main tool of the extreme values approach: extrapolation over the unit of the return period.
The first thing that must be defined is: what is the unit of the return period? In this example, the unit is days because the observations are daily, but in other applications, like corrosion engineering, the unit may be number of observations.
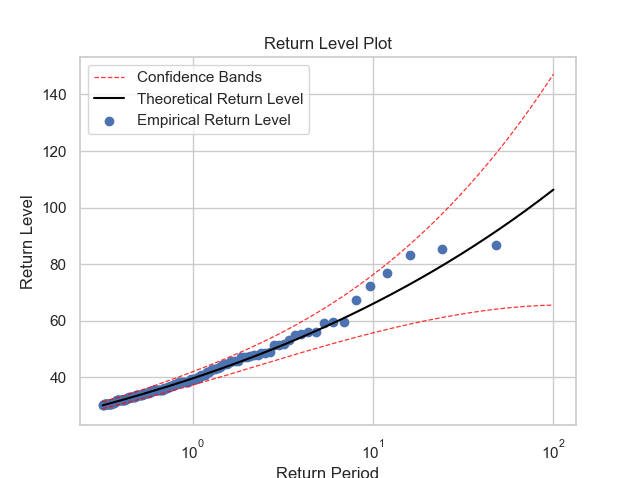
Using the function return_value is possible to get two informations:
- 1 : The return value for a given return period and;
- 2 : The return value plot, that works very well for a model diagnostic.
By running this line (go to Model Diagnostics and Return Level Analysis for more information about the function):
thresh_modeling.return_value(data, 30, 0.05, 365, 36500, 'mle')It means, the return period we want to know the exact return value is 36500 days or 100 years. With the 365, we are saying that the annual number of observations is 365.
The results must be:
The return value for the given return period is 106.34386649996667 ± 40.86691363790978
Hence, by the graphic, it is possible to say that the theoretical model is very well fitted. Also, it was possible to compute the return value in 100 years. In other words, the rainfall preciptation once in every 100 years must be between 65.4470 and 147.2108 mm.
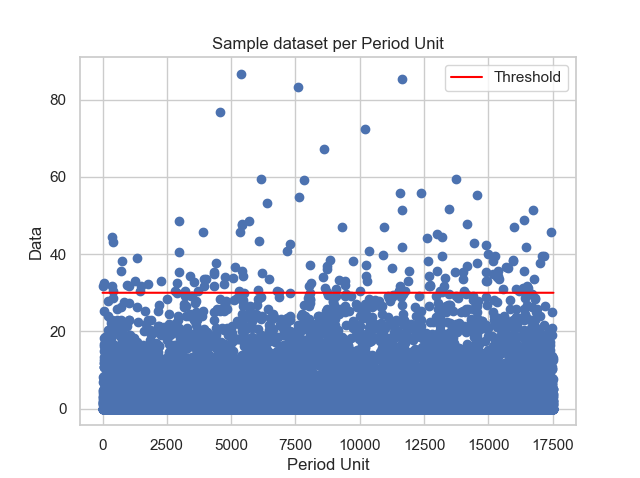
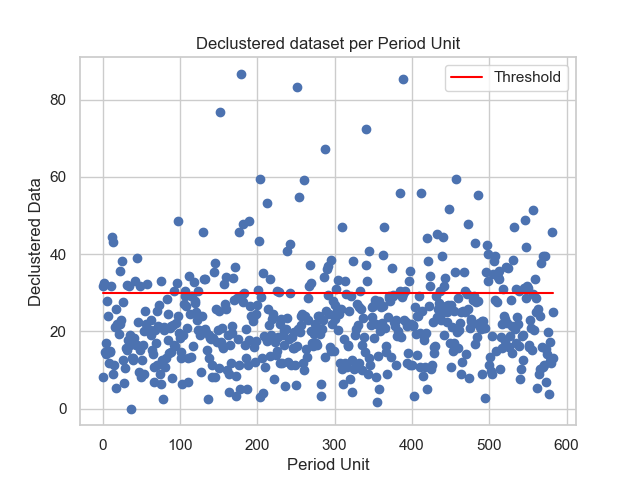
Stuart Coles's in his book says that if the extremes assume a tendency to be clustered in a stationary series, another pratice would be need to model these values. The pratice consists in declustering, which is: cluster data and decluster by its maximums. For this example, it is clear that, at least initialy, the dataset is not orgnanized in clusters. With the function decluster it is possible to observe the dataset plot against its unit of return period, but, also it is possible to cluster it using a given block size (in this example it will be monthly, then the block size will be 30 days), and then decluster it by taking the maximum of each block.
By running these lines:
thresh_modeling.decluster(data, 30, 30)The result must be:
It is important to say that the unit of the return period after the decluster changes (monthly). With the first graph is possible to observe that, at least initialy, there is not any pattern. However, it does not means that it is not possible to descluter the data set to a given block size, which is possible to see in the second graphic.
In a case that it is necessary to decluster the dataset, the second one, shown in the declustered graphic must be used.
The other functions that are not in this tutorial can be used as it is shown in the test file. The discription of each one is in the Functions Documenation.
Any doubts about the package, don't hesitate to contact me.
Copyright (c) 2019 Iago Pereira Lemos
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see https://www.gnu.org/licenses/
For referencing the repository, use the following code:
@misc{thresholdmodeling,
author = {Iago P. Lemos and Antonio Marcos G. Lima and Marcus Antonio Viana Duarte},
title = {thresholdmodeling package},
month = Feb,
year = 2020,
doi = {10.5281/zenodo.3661338},
version = {0.0.1},
publisher = {Zenodo},
url = {https://github.com/iagolemos1/thresholdmodeling}
}
I am a mechanical engineering undergraduate student in the Federal University of Uberlândia and this package was made in the Acoustics and Vibration Laboratory, in the School of Mechanical Engineering.