DHSort is a convolutional neural network (CNN) develop to classify haploid maize seeds based on seed images obtained from induction progenies using haploid inducers equipped with the R1-nj marker.
More details about the development of DHSort are available on Improving the identification of haploid maize seeds using Convolutional Neural Networks.
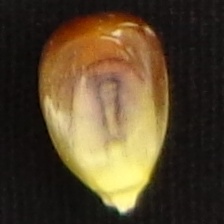
Doubled haploid (DH) technology is employed to accelerate the development of new maize lines. Briefly, the DH technique consists of obtaining haploids individuals, identification of haploid within induction crosses, and an artificial chromosomal doubling of haploid seedlings. This method permits shorten the time used in this stage. In maize, the principal method applied to obtain haploid individuals is using an in vivo haploid inducer. These inducers are genotypes that have a high rate of haploid individuals in their progenies. Overall, seeds obtained from induction crosses can be haploids or diploids, and the main system used to sort them is the R1-nj marker, according to images below:
| diploid (D) | haploid (H) | inhibited (I) |
|---|---|---|
 |
 |
 |
However, the anthocyanin expressiveness is variable, which makes it difficult to distinguish between seed classes. We developed a CNN model to classify accurately haploid maize seeds using R1-nj avoiding sorting mistakes.
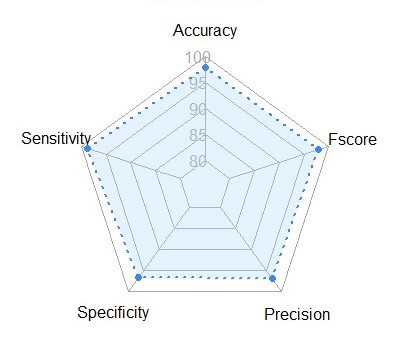
DHSort was highly accurate for maize haploid seed sorting task. Below are the performance metrics:
|
 |
First of all, you should install the following software and packages to use DHSort:
- Python 3 (https://www.python.org/)
- Tensorflow (https://www.tensorflow.org/)
- Keras (https://keras.io/)
- numpy
- pandas
1. Download the repository by clicking on the button or type the following command in CMD to clone the repository
git clone https://github.com/sabadinfelipe/DHsort.git
2. Into dataset directory, there are 60 seed image examples (30 haploids and 30 diploida). To predict them, run the following code:
python DHsort.py
3. Results are stored in an output.csv file, which contains two columns: file_name and phenotype. The file_name column is composed of image filenames, whereas the phenotype column is its prediction. D refers to diploid and H refers to a haploid seed. Below, it follows an output example:
| file_name | phenotype |
|---|---|
| diploid_1.jpg | D |
| haploid_5.jpg | H |
| diploid_2.jpg | D |
| diploid_10.jpg | D |
If you want to predict your seed images, delete image files into the datasetdirectory, and move your image dataset to it. After, run the DHsort.py script to predict them. It will create an output.csv file containing the predictions. Attention: Seed images should look like to Figure 1 (only one seed centered and a black background).
Figure 1. Seed image example for input in the DHSort.
Sabadin, F., Galli, G., Borsato, R., Jr, Gevartosky, R., Campos, G.R. and Fritsche‐Neto, R. (2021), Improving the identification of haploid maize seeds using convolutional neural networks. Crop Science. https://doi.org/10.1002/csc2.20487
I would like to thank Allogamous Plant Breeding Laboratory team for supporting in this project.
Please report any question or bug to this email.
Allogamous Plant Breeding Lab, University of São Paulo - Brazil