Our project aims to give a software solution to the problem of accidental neoplasm findings in CT scans – especially emergency medicine. Using said software, CT scan slices would be autonomously processed and analysed to screen the data and mark the slices with suspicious abnormalities. Those slices would then be reviewed by a radiologist, who would give the final verdict on the possible malignancy of the abnormality.
In practice, a successful, large scale screening method could significantly increase the cancer survival rate as its prognosis is greatly dependent on the stage said cancer is in at the time of the diagnosis.
This project was done at the JunctionX Budapest 48-hour hackathon and is our approach to the challenge given by Varian, which can be found here.
In the given challenge we are focusing solely on brain cancer CT scans, but similar technology could be applied to any CT/MRI scan.
A raw, unlabeled, dataset of CT and MR DICOM images for 30 patients was provided. After examining the challenge, we decided for two different approaches to the problem:
- Deep Learning Approach
- Brain Symmetry Approach
For supervised learning, labelled data is a requirement. Since the provided data wasn't labelled, we came up with a method to properly label it. For each series of scans, there is a file that indicates and contours the tumor(s) on each abnormal slice. We, therefore, decided to use that information to label each of the slices as either containing tumor or not containing tumor, depending on the tumor's presence.
The next step was to decide which model architecture we should use. Due to strict time limitation, we couldn't dive too deep into the problem and decided to go with pretrained VGG16 on Imagenet. Unfortunately, as expected, the model overfits on our dataset due to its small quantity and uneven distribution (there being more slices not containing any tumors than those that do). The second issue can be easily eliminated by solving the first one (using a larger dataset), which is something we are looking to deal with in the near future.
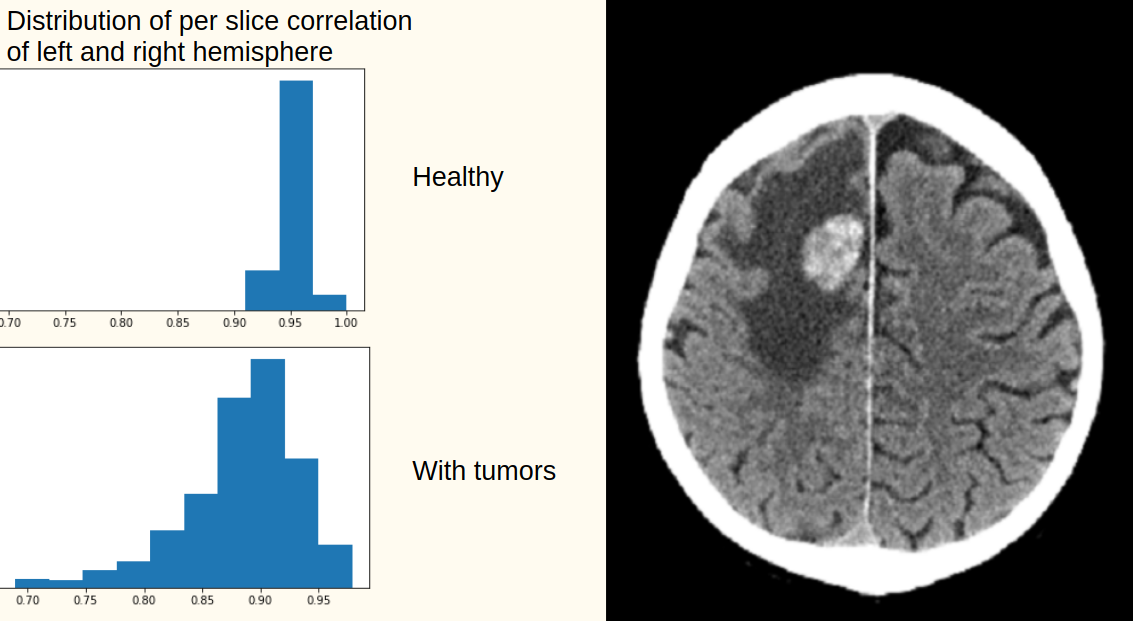
Given very limited amount of data we decided to take advantage of the natural symmetry of the right and left brain hemisphere. Healthy brains exhibit high correlation between hemispheres (over 0.9, see the figure below), whereas a potential brain tumor breaks this symmetry. This approach requires perfect orientation of the brain, meaning no rotation in either of the axis and the middle line cuts the brain to two hemispheres. We search this orientation with a random walk over different orientations and middle lines to maximise the slice symmetry.
Slices around the middle (of the vertical axis) with correlations 0.8 to 0.9 are more likely to contain tumor. Slices around the edges (the top of the brain, and the bottom) contains very little part of brain or bones which yields very low and unpredictable correlations and thus the tumor cannot be detected in this regions with the symmetry technique.