nf-core/smartseq
Please use the version on nf-core: https://github.com/nf-core/smartseq2
Preprocess scRNA-seq data generated with the SmartSeq2 protocol..
Introduction
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It comes with docker containers making installation trivial and results highly reproducible.
Documentation
The nf-core/smartseq pipeline comes with documentation about the pipeline, found in the docs/ directory:
- Installation
- Pipeline configuration
- Running the pipeline
- Output and how to interpret the results
- Troubleshooting
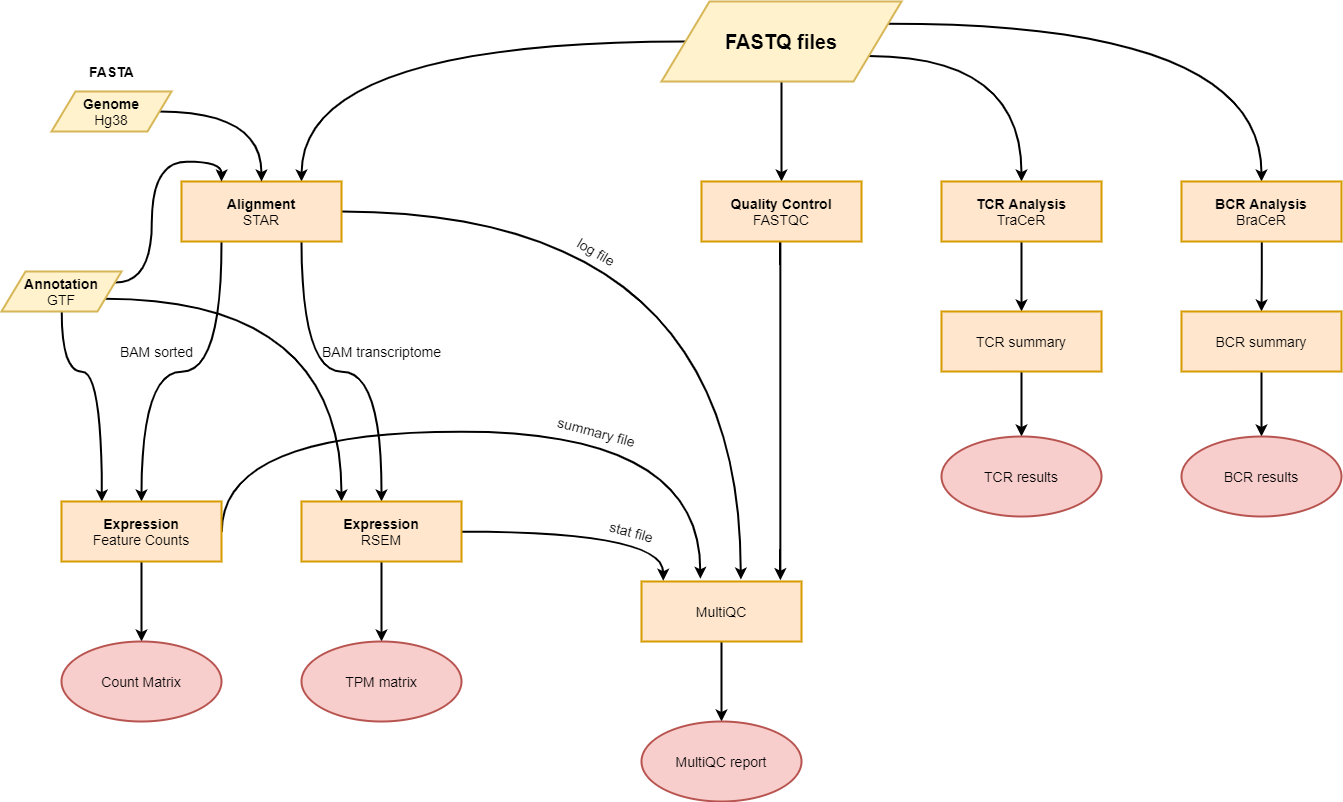
This pipeline was developed to process single cell RNA-seq data generated using the Smart-Seq2 protocol.
The pipeline features:
- Quality control using
fastqcandmultiqc - Alignment using
STAR - Quantification using
rsemorfeatureCounts - TCR analysis using
TraCeR. - BCR analysis using
BraCeR.
Credits
nf-core/smartseq was originally written by Sandro Carollo, Giorgos Fotakis and Gregor Sturm