This repo is part of web/apps for Final Projects BANGKIT ACADEMY 2020 lead by Google, Gojek, Tokopedia, and Traveloka | BDG2-A
For more information that can find in the GitHub link: https://github.com/ilmimris/bangkit-bgd2a-webapp
How to predict images from CT-Scan and X-Ray Chest infected Coronavirus or not?
* A success metric is that images can predict what CT-Scan or X-Ray Chest infected Coronavirus or not.
* Success means the numbers of predicting upper 94% accuracy from that images.
* Failure means the result of predicting no better than heuristics.
* Consider collected images from the dataset that categories Covid19 or not in the past. Assume that images will labialize that Covid19 image or not.
* Image recognition using Transfer Learning with model sequential with using Covid Chestxray Dataset & Chest X-Ray From Kaggle
* Image recognition (alternative model) using Transfer Learning with model sequential with using Dataset Repository Mosmed COVID-19 CT Scans from Kaggle & COVID-CT from GitHub
https://github.com/ieee8023/covid-chestxray-dataset
🛑 Note: please do not claim diagnostic performance of a model without a clinical study! This is not a kaggle competition dataset. Please read this paper about evaluation issues: https://arxiv.org/abs/2004.12823 and https://arxiv.org/abs/2004.05405
Project Summary: To build a public open dataset of chest X-ray and CT images of patients which are positive or suspected of COVID-19 or other viral and bacterial pneumonias (MERS, SARS, and ARDS.). Data will be collected from public sources as well as through indirect collection from hospitals and physicians. All images and data will be released publicly in this GitHub repo.
This project is approved by the University of Montreal's Ethics Committee #CERSES-20-058-D
Current stats of PA, AP, and AP Supine views. Labels 0=No or 1=Yes.
COVID19_Dataset num_samples=408 views=['PA', 'AP']
{'ARDS': {0.0: 393, 1.0: 15},
'Bacterial Pneumonia': {0.0: 379, 1.0: 29},
'COVID-19': {0.0: 96, 1.0: 312},
'Chlamydophila': {0.0: 407, 1.0: 1},
'Fungal Pneumonia': {0.0: 395, 1.0: 13},
'Influenza': {0.0: 407, 1.0: 1},
'Klebsiella': {0.0: 401, 1.0: 7},
'Legionella': {0.0: 404, 1.0: 4},
'Lipoid': {0.0: 405, 1.0: 3},
'MERS': {0.0: 408},
'Mycoplasma': {0.0: 404, 1.0: 4},
'No Finding': {0.0: 393, 1.0: 15},
'Pneumocystis': {0.0: 395, 1.0: 13},
'Pneumonia': {0.0: 17, 1.0: 391},
'SARS': {0.0: 392, 1.0: 16},
'Streptococcus': {0.0: 395, 1.0: 13},
'Varicella': {0.0: 404, 1.0: 4},
'Viral Pneumonia': {0.0: 75, 1.0: 333}}
COVID19_Dataset num_samples=134 views=['AP Supine']
{'ARDS': {0.0: 133, 1.0: 1},
'Bacterial Pneumonia': {0.0: 134},
'COVID-19': {0.0: 12, 1.0: 122},
'Chlamydophila': {0.0: 134},
'Fungal Pneumonia': {0.0: 134},
'Influenza': {0.0: 134},
'Klebsiella': {0.0: 134},
'Legionella': {0.0: 134},
'Lipoid': {0.0: 134},
'MERS': {0.0: 134},
'Mycoplasma': {0.0: 134},
'No Finding': {0.0: 130, 1.0: 4},
'Pneumocystis': {0.0: 134},
'Pneumonia': {0.0: 8, 1.0: 126},
'SARS': {0.0: 134},
'Streptococcus': {0.0: 134},
'Varicella': {0.0: 134},
'Viral Pneumonia': {0.0: 12, 1.0: 122}}
https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
http://www.cell.com/cell/fulltext/S0092-8674(18)30154-5
The normal chest X-ray (left panel) depicts clear lungs without any areas of abnormal opacification in the image. Bacterial pneumonia (middle) typically exhibits a focal lobar consolidation, in this case in the right upper lobe (white arrows), whereas viral pneumonia (right) manifests with a more diffuse ‘‘interstitial’’ pattern in both lungs. http://www.cell.com/cell/fulltext/S0092-8674(18)30154-5
The dataset is organized into 3 folders (train, test, val) and contains subfolders for each image category (Pneumonia/Normal). There are 5,863 X-Ray images (JPEG) and 2 categories (Pneumonia/Normal).
Chest X-ray images (anterior-posterior) were selected from retrospective cohorts of pediatric patients of one to five years old from Guangzhou Women and Children’s Medical Center, Guangzhou. All chest X-ray imaging was performed as part of patients’ routine clinical care.
For the analysis of chest x-ray images, all chest radiographs were initially screened for quality control by removing all low quality or unreadable scans. The diagnoses for the images were then graded by two expert physicians before being cleared for training the AI system. In order to account for any grading errors, the evaluation set was also checked by a third expert.
- Data: https://data.mendeley.com/datasets/rscbjbr9sj/2
- License: CC BY 4.0
- Citation: http://www.cell.com/cell/fulltext/S0092-8674(18)30154-5
Usage Information License Other (specified in description)
Visibility Public
Maintainers Dataset owner Paul Mooney(Kaggle)
Updates Expected update frequency Not specified
Last updated 2018-03-25
Date created 2018-03-22
Current version Version 2
https://www.kaggle.com/andrewmvd/mosmed-covid19-ct-scans
This dataset contains anonymised human lung computed tomography (CT) scans with COVID-19 related findings, as well as without such findings. In total, there are 1000 CT scans each from a unique patient.
A subset of 50 studies has been annotated with binary pixel masks for segmentation depicting regions of interest (ground-glass opacifications and consolidations). CT scans were obtained between 1st of March, 2020 and 25th of April, 2020, and provided by medical hospitals in Moscow, Russia.
If you use this dataset in your research, please credit the authors
Morozov, S., Andreychenko, A., Blokhin, I., Vladzymyrskyy, A., Gelezhe, P., Gombolevskiy, V., Gonchar, A., Ledikhova, N., Pavlov, N., Chernina, V. MosMedData: Chest CT Scans with COVID-19 Related Findings, 2020, v. 1.0, link
CC BY NC ND 3.0
Image by rawpixel
Usage Information License Other (specified in description)
Visibility Public
Provenance Sources soon
Collection methodology Please refer to description and sources
Maintainers Dataset owner Larxel(Kaggle)
Updates Expected update frequency Never
Last updated 2020-05-25
Date created 2020-05-25
Current version Version 5
https://github.com/UCSD-AI4H/COVID-CT.git
The utility of this dataset has been confirmed by a senior radiologist in Tongji Hospital, Wuhan, China, who has performed diagnosis and treatment of a large number of COVID-19 patients during the outbreak of this disease between January and April.
After releasing this dataset, we received several feedback expressing concerns about the usability of this dataset. The major concerns are summarized as follows. First, when the original CT images are put into papers, the quality of these images are degraded, which may render the diagnosis decisions less accurate. The quality degradation includes: the Hounsfield unit (HU) values are lost; the number of bits per pixel is reduced; the resolution of images is reduced. Second, the original CT scan contains a sequence of CT slices, but when put into papers, only a few key slices are selected, which may have negative impact on diagnosis as well.
We consulted the aforementioned radiologist at Tongji Hospital regarding these two concerns. According to the radiologist, the issues raised in these concerns do not significantly affect the accuracy of diagnosis decision-making. First, experienced radiologists are able to make accurate diagnosis from low quality CT images. For example, given a photo taken by smart phone of the original CT image, experienced radiologists can make accurate diagnosis by just looking at the photo, though the CT image in the photo has much lower quality than the original CT image. Likewise, the quality gap between CT images in papers and original CT images will not largely hurt the accuracy of diagnosis. Second, while it is preferable to read a sequence of CT slices, oftentimes a single-slice of CT contains enough clinical information for accurate decision-making.
The COVID-CT-Dataset has 349 CT images containing clinical findings of COVID-19 from 216 patients. They are in ./Images-processed/CT_COVID.zip
Non-COVID CT scans are in ./Images-processed/CT_NonCOVID.zip
We provide a data split in ./Data-split. Data split information see README for DenseNet_predict.md
The meta information (e.g., patient ID, patient information, DOI, image caption) is in COVID-CT-MetaInfo.xlsx
The images are collected from COVID19-related papers from medRxiv, bioRxiv, NEJM, JAMA, Lancet, etc. CTs containing COVID-19 abnormalities are selected by reading the figure captions in the papers. All copyrights of the data belong to the authors and publishers of these papers.
The dataset details are described in this preprint: COVID-CT-Dataset: A CT Scan Dataset about COVID-19
If you find this dataset and code useful, please cite:
@article{zhao2020COVID-CT-Dataset,
title={COVID-CT-Dataset: a CT scan dataset about COVID-19},
author={Zhao, Jinyu and Zhang, Yichen and He, Xuehai and Xie, Pengtao},
journal={arXiv preprint arXiv:2003.13865},
year={2020}
}
We developed two baseline methods for the community to benchmark with. The code are in the "baseline methods" folder and the details are in the readme files under that folder. The methods are described in Sample-Efficient Deep Learning for COVID-19 Diagnosis Based on CT Scans
- To contribute to our project, please email your data to jiz077@eng.ucsd.edu with the corresponding meta information (Patient ID, DOI and Captions).
- We recommend you also extract images from publications or preprints. Make sure the original papers you crawled have different DOIs from those listed in COVID-CT-MetaInfo.xlsx.
- In COVID-CT-MetaInfo.xlsx, images with the form of 2020.mm.dd.xxxx are crawled from bioRxiv or medRxiv. The DOIs for these preprints are 10.1101/2020.mm.dd.xxxx.
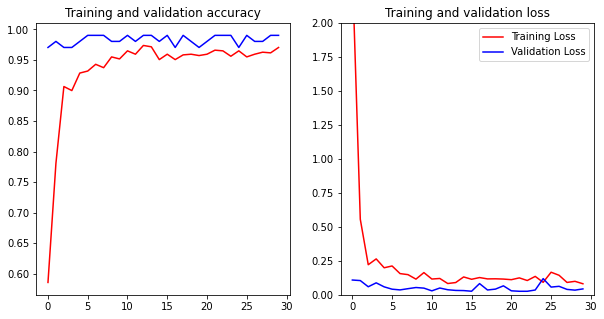
The Accuracy and Validation accuracy reaches 97% and 99%. That means the models have success metrics.
The graph titled "Training and Validation Accuracy" that could see, the distance of training data and validation is close so that overfitting does not occur.