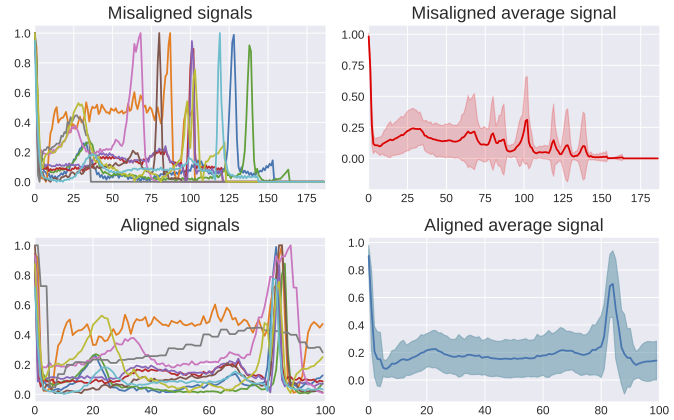
Employing differentiable alignment to improve ECG classification.
Place data file train.csv and test.csv under the ./data/ folder.
Install the corresponding dependencies:
pip install requirements.txtTo run the model, using the
python main.py --model {model} \
--log_path {log_path} \
--ckpt_path {log_path} \
--epoch {epoch_num} \
--lr {learning_rate} \
--batch_size {batch_size} \
--seed {random_seed} \
--align {align_point_len} \
--inference
-
model: The backbone model, select from[grufcn, trans, fcn, resnet, rescnn, gruatt, tcn] -
align(opt.): The length of the aligned signal. If not set, no alignment is performed. -
inference(opt.): Conduct inference over./data/test.csv.For more details, please refer to
run.shandmain.py.
Part of the codes is reused from tsai and DTAN.
Our code in this repository is licensed under the MIT license.